8V17
 
 | |
8GCY
 
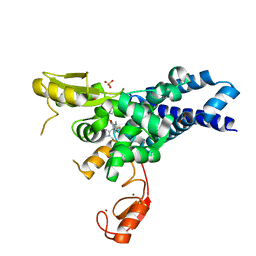 | | Co-crystal structure of CBL-B in complex with N-Aryl isoindolin-1-one inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-{3-[(1s,3R)-3-methyl-1-(4-methyl-4H-1,2,4-triazol-3-yl)cyclobutyl]phenyl}-6-{[(3S)-3-methylpiperidin-1-yl]methyl}-4-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, E3 ubiquitin-protein ligase CBL-B, ... | | Authors: | Kimani, S, Zeng, H, Dong, A, Li, Y, Santhakumar, V, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-03 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The co-crystal structure of Cbl-b and a small-molecule inhibitor reveals the mechanism of Cbl-b inhibition.
Commun Biol, 6, 2023
|
|
8BK0
 
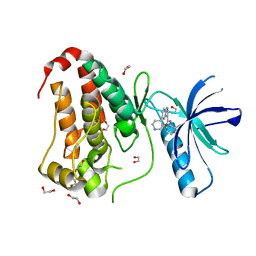 | | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904 | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, ~{N}-(2-chlorophenyl)-6-piperidin-4-yl-imidazo[1,2-a]pyridine-3-carboxamide | | Authors: | Zhubi, R, Gerninghaus, J, Knapp, S, Kraemer, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human Ephrin type-A receptor 2 (EPHA2) Kinase domain in complex with LDN-211904
To Be Published
|
|
8VB5
 
 | |
6UEH
 
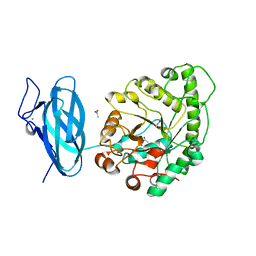 | | Crystal structure of a ruminal GH26 endo-beta-1,4-mannanase | | Descriptor: | ACETATE ION, CALCIUM ION, Cow rumen GH26 endo-mannanase | | Authors: | Mandelli, F, Morais, M.A.B, Lima, E.A, Persinoti, G.F, Murakami, M.T. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
J.Biol.Chem., 295, 2020
|
|
8BSR
 
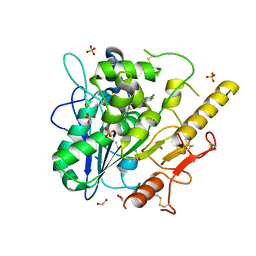 | | Notum Inhibitor ARUK3006562 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-26 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
8T93
 
 | | Crystal structure of Terrestrivirus Inositol pyrophosphatase kinase in complex with ADP and scyllo-L-(1,2,3,4)-IP4 | | Descriptor: | (1R,2S,3S,4R,5S,6S)-5,6-dihydroxycyclohexane-1,2,3,4-tetrayl tetrakis[dihydrogen (phosphate)], ADENOSINE-5'-DIPHOSPHATE, Kinase, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2023-06-23 | | Release date: | 2023-11-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an inositol pyrophosphate kinase from a giant virus.
Embo J., 43, 2024
|
|
8BTE
 
 | | Notum Inhibitor ARUK3004470 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2022-11-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Designed switch from covalent to non-covalent inhibitors of carboxylesterase Notum activity.
Eur.J.Med.Chem., 251, 2023
|
|
4RYL
 
 | | Human Protein Arginine Methyltransferase 3 in complex with 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea | | Descriptor: | 1-isoquinolin-6-yl-3-[2-oxo-2-(pyrrolidin-1-yl)ethyl]urea, PRMT3 protein, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Dobrovetsky, E, Kaniskan, H.U, Szewczyk, M, Yu, Z, Eram, M.S, Yang, X, Schmidt, K, Luo, X, Dai, M, He, F, Zang, I, Lin, Y, Kennedy, S, Li, F, Tempel, W, Smil, D, Min, S.J, Landon, M, Lin-Jones, J, Huang, X.P, Roth, B.L, Schapira, M, Atadja, P, Barsyte-Lovejoy, D, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Brown, P.J, Zhao, K, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-12-15 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Potent, Selective and Cell-Active Allosteric Inhibitor of Protein Arginine Methyltransferase 3 (PRMT3).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
8DWK
 
 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
8PNO
 
 | |
1E7R
 
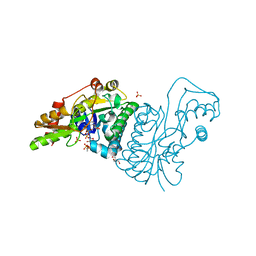 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
4TTE
 
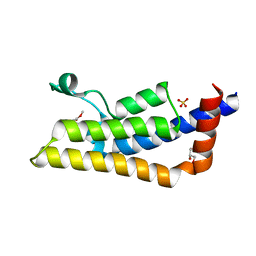 | | Crystal structure of ATAD2A bromodomain complexed with methyl 3-amino-5-(3,5-dimethyl-1,2-oxazol-4-yl)benzoate | | Descriptor: | ATPase family AAA domain-containing protein 2, CHLORIDE ION, GLYCEROL, ... | | Authors: | Poncet-Montange, G, Zhan, Y, Bardenhagen, J, Petrocchi, A, Leo, E, Shi, X, Lee, G, Leonard, P, Geck Do, M, Cardozo, M, Palmer, W, Andersen, J, Jones, P, Ladbury, J. | | Deposit date: | 2014-06-20 | | Release date: | 2014-12-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Observed bromodomain flexibility reveals histone peptide- and small molecule ligand-compatible forms of ATAD2.
Biochem.J., 466, 2015
|
|
6XDH
 
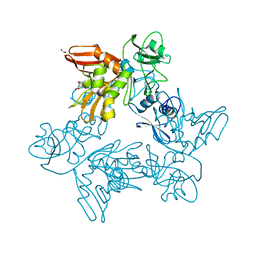 | |
8VKZ
 
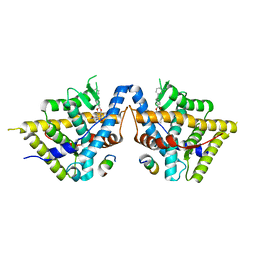 | | Crystal structure of Glucocorticoid Receptor in complex with an inhibitor | | Descriptor: | (4aR,4bS,5R,6aS,6bS,8R,9aR,10aR,10bR)-8-{4-[(3-aminophenyl)methyl]phenyl}-5-hydroxy-6b-(hydroxyacetyl)-4a,6a-dimethyl-4a,4b,5,6,6a,6b,9a,10,10a,10b,11,12-dodecahydro-2H,8H-naphtho[2',1':4,5]indeno[1,2-d][1,3]dioxol-2-one, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2024-01-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Minimising the payload solvent exposed hydrophobic surface area optimises the antibody-drug conjugate properties.
Rsc Med Chem, 15, 2024
|
|
7SFM
 
 | | Mycobacterium tuberculosis Hip1 crystal structure | | Descriptor: | ACETATE ION, GLYCEROL, Hip1 | | Authors: | Ostrov, D.A, Li, D. | | Deposit date: | 2021-10-04 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | 2.1 angstrom crystal structure of the Mycobacterium tuberculosis serine hydrolase, Hip1, in its anhydro-form (Anhydrohip1).
Biochem.Biophys.Res.Commun., 630, 2022
|
|
9EP6
 
 | | Alpha-Galactosaminidase family GH114 from Fusarium solani | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, alpha-galactosidase | | Authors: | Roth, C, Moroz, O.V, Miranda, S.A.D, Jahn, L, Blagova, E, Lebedev, A.A, Segura, D.R, Stringer, M.A, Friis, E.P, Davies, G.J, Wilson, K.S. | | Deposit date: | 2024-03-17 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structures of alpha-galactosaminidases from the CAZy GH114 family and homologs defining a new GH191 family of glycosidases.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
6BJT
 
 | | The structure of AtzH: a little known member of the atrazine breakdown pathway | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peat, T.S, Newman, J, Scott, C, Esquirol, L. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel decarboxylating amidohydrolase involved in avoiding metabolic dead ends during cyanuric acid catabolism in Pseudomonas sp. strain ADP.
PLoS ONE, 13, 2018
|
|
6FH2
 
 | | Protein arginine kinase McsB in the AMP-PN-bound state | | Descriptor: | 1,2-ETHANEDIOL, AMP PHOSPHORAMIDATE, Protein-arginine kinase | | Authors: | Suskiewicz, M.J, Heuck, A, Vu, L.D, Clausen, T. | | Deposit date: | 2018-01-12 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of McsB, a protein kinase for regulated arginine phosphorylation.
Nat.Chem.Biol., 15, 2019
|
|
9J1E
 
 | |
1EXV
 
 | | HUMAN LIVER GLYCOGEN PHOSPHORYLASE A COMPLEXED WITH GLCNAC AND CP-403,700 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LIVER GLYCOGEN PHOSPHORYLASE, N-acetyl-beta-D-glucopyranosylamine, ... | | Authors: | Rath, V.L, Ammirati, M, Danley, D.E, Ekstrom, J.L, Hynes, T.R, Olson, T.V, Hoover, D.J. | | Deposit date: | 2000-05-04 | | Release date: | 2000-11-01 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human liver glycogen phosphorylase inhibitors bind at a new allosteric site.
Chem.Biol., 7, 2000
|
|
8GNK
 
 | | CryoEM structure of cytosol-facing, substrate-bound ratGAT1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Nayak, S.R, Joseph, D, Penmatsa, A. | | Deposit date: | 2022-08-24 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structure of GABA transporter 1 reveals substrate recognition and transport mechanism.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GHV
 
 | | Cannabinoid Receptor 1-G Protein Complex | | Descriptor: | (5Z,8Z,11Z,13S,14Z)-N-[(2R)-1-hydroxypropan-2-yl]-13-methylicosa-5,8,11,14-tetraenamide, Cannabinoid receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Krishna Kumar, K, Robertson, M.J, Skiniotis, G, Kobilka, B.K. | | Deposit date: | 2023-03-12 | | Release date: | 2023-05-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for activation of CB1 by an endocannabinoid analog.
Nat Commun, 14, 2023
|
|
8GB8
 
 | |
6PL9
 
 | | Adduct formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidaz ol-2-ylidene)(eta5-pentamethylcyclopentadienyl)rhodium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-rhodapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
