4M1M
 
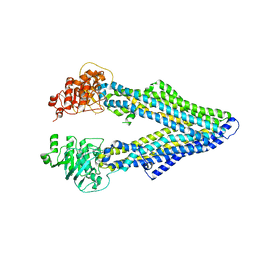 | |
6D9N
 
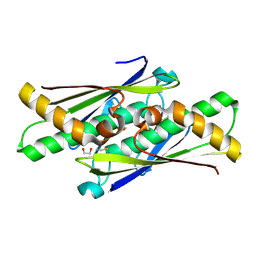 | |
6GDI
 
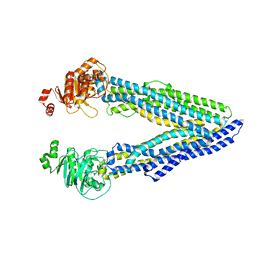 | | Structure of P-glycoprotein(ABCB1) in the post-hydrolytic state | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ford, R.C, Thonghin, N, Collins, R.F, Barbieri, A, Shafi, T, Siebert, A. | | Deposit date: | 2018-04-23 | | Release date: | 2018-05-23 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Novel features in the structure of P-glycoprotein (ABCB1) in the post-hydrolytic state as determined at 7.9 angstrom resolution.
Bmc Struct.Biol., 18, 2018
|
|
4LSG
 
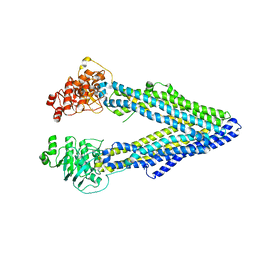 | |
4Q9H
 
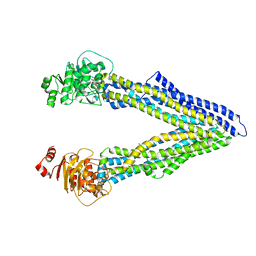 | | P-glycoprotein at 3.4 A resolution | | Descriptor: | Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9J
 
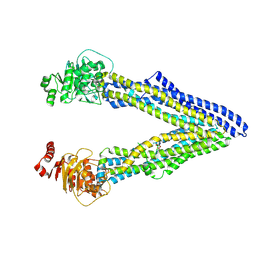 | | P-glycoprotein cocrystallised with QZ-Val | | Descriptor: | Multidrug resistance protein 1A, N-{[5-AMINO-1-(5-O-PHOSPHONO-BETA-D-ARABINOFURANOSYL)-1H-IMIDAZOL-4-YL]CARBONYL}-L-ASPARTIC ACID | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9K
 
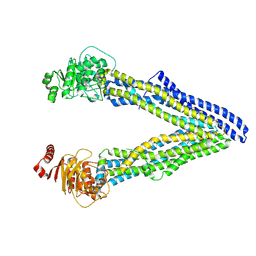 | | P-glycoprotein cocrystallised with QZ-Leu | | Descriptor: | (30F)L(30F)L(30F)L Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4Q9I
 
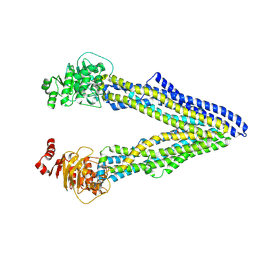 | | P-glycoprotein cocrystallised with QZ-Ala | | Descriptor: | (30F)A(30F)A(30F)A Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.781 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XWK
 
 | | P-glycoprotein co-crystallized with BDE-100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Rees, S.D, Chang, G. | | Deposit date: | 2015-01-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure.
Sci Adv, 2, 2016
|
|
4Q9L
 
 | | P-glycoprotein cocrystallised with QZ-Phe | | Descriptor: | (30F)F(30F)F(30F)F Peptide, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Szewczyk, P, Chang, G. | | Deposit date: | 2014-05-01 | | Release date: | 2015-03-04 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Snapshots of ligand entry, malleable binding and induced helical movement in P-glycoprotein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4M2S
 
 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-RRR | | Descriptor: | (4R,11R,18R)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
4M2T
 
 | | Corrected Structure of Mouse P-glycoprotein bound to QZ59-SSS | | Descriptor: | (4S,11S,18S)-4,11,18-tri(propan-2-yl)-6,13,20-triselena-3,10,17,22,23,24-hexaazatetracyclo[17.2.1.1~5,8~.1~12,15~]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, Multidrug resistance protein 1A | | Authors: | Li, J, Jaimes, K.F, Aller, S.G. | | Deposit date: | 2013-08-05 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.35 Å) | | Cite: | Refined structures of mouse P-glycoprotein.
Protein Sci., 23, 2014
|
|
6C0V
 
 | |
8WXO
 
 | |
8WXN
 
 | |
8WXM
 
 | |
8WXP
 
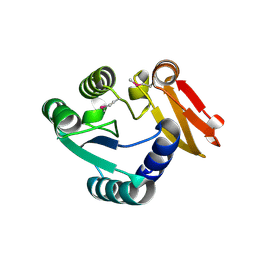 | |
2XA6
 
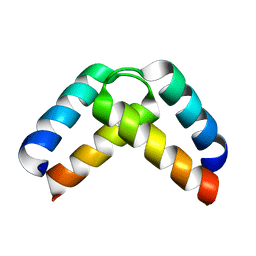 | | Structural basis for homodimerization of the Src-associated during mitosis, 68 kD protein (Sam68) Qua1 domain | | Descriptor: | KH DOMAIN-CONTAINING,RNA-BINDING,SIGNAL TRANSDUCTION-ASSOCIATED PROTEIN 1 | | Authors: | Meyer, N.H, Tripsianes, K, Vincendeaux, M, Madl, T, Kateb, F, Brack-Werner, R, Sattler, M. | | Deposit date: | 2010-03-29 | | Release date: | 2010-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Structural basis for homodimerization of the Src-associated during mitosis, 68-kDa protein (Sam68) Qua1 domain.
J. Biol. Chem., 285, 2010
|
|
7Z9A
 
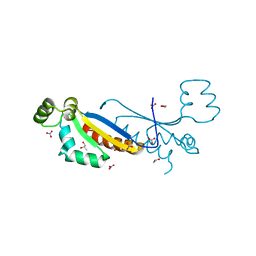 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, Isoform 2 of KH domain-containing, RNA-binding, ... | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-20 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
7Z89
 
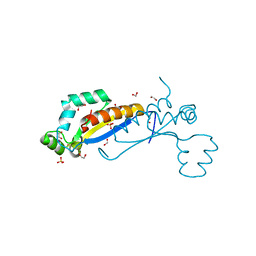 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Isoform 2 of KH domain-containing, ... | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
7ZAB
 
 | | Sam68 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nadal, M, Puestes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
7ZAM
 
 | | Sam68 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, IODIDE ION, ... | | Authors: | Nadal, M, Fuentes-Prior, P. | | Deposit date: | 2022-03-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structure and function analysis of Sam68 and hnRNP A1 synergy in the exclusion of exon 7 from SMN2 transcripts.
Protein Sci., 32, 2023
|
|
4DR2
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DR4
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, cognate transfer RNA anticodon stem-loop and multiple copies of paromomycin molecules bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.969 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
4DR7
 
 | | Crystal structure of the Thermus thermophilus (HB8) 30S ribosomal subunit with codon, crystallographically disordered near-cognate transfer RNA anticodon stem-loop mismatched at the second codon position, and streptomycin bound | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2012-02-16 | | Release date: | 2012-11-14 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | A structural basis for streptomycin-induced misreading of the genetic code.
Nat Commun, 4, 2013
|
|
