5K7U
 
 | |
5K8O
 
 | | Mn2+/5NSA-bound 5-nitroanthranilate aminohydrolase | | Descriptor: | 5-nitroanthranilic acid aminohydrolase, 5-nitrosalicylic acid, MANGANESE (II) ION | | Authors: | Kalyoncu, S. | | Deposit date: | 2016-05-30 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.893 Å) | | Cite: | Enzymatic hydrolysis by transition-metal-dependent nucleophilic aromatic substitution.
Nat.Chem.Biol., 12, 2016
|
|
5JYX
 
 | |
5K02
 
 | | Structure of human SOD1 with T2D mutation | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Fay, J.M, Zhu, C, Cui, W, Ke, H, Dokholyan, N.V. | | Deposit date: | 2016-05-17 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Phosphomimetic Mutation Stabilizes SOD1 and Rescues Cell Viability in the Context of an ALS-Associated Mutation.
Structure, 24, 2016
|
|
5KBA
 
 | | Computational Design of Self-Assembling Cyclic Protein Homooligomers | | Descriptor: | Designed protein ANK1C2 | | Authors: | Sankaran, B, Zwart, P.H, Fallas, J.A, Pereira, J.H, Ueda, G, Baker, D. | | Deposit date: | 2016-06-02 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Computational design of self-assembling cyclic protein homo-oligomers.
Nat Chem, 9, 2017
|
|
5KBX
 
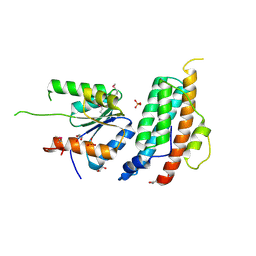 | |
5JQ9
 
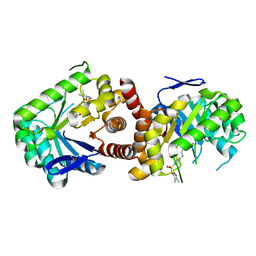 | | Yersinia pestis DHPS with pterine-sulfa conjugate Compound 16 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2-amino-4-oxo-3,4-dihydropteridin-6-yl)methyl]amino}-N-(3,4-dimethyl-1,2-oxazol-5-yl)benzene-1-sulfonamide, Dihydropteroate synthase | | Authors: | Wu, Y, White, S.W. | | Deposit date: | 2016-05-04 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Pterin-sulfa conjugates as dihydropteroate synthase inhibitors and antibacterial agents.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5JTW
 
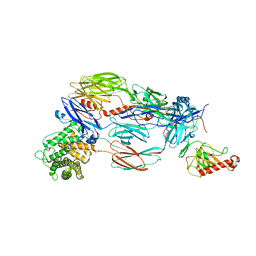 | | Crystal structure of complement C4b re-refined using iMDFF | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C4-A | | Authors: | Croll, T.I, Andersen, G.R. | | Deposit date: | 2016-05-09 | | Release date: | 2016-08-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Re-evaluation of low-resolution crystal structures via interactive molecular-dynamics flexible fitting (iMDFF): a case study in complement C4.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5KN9
 
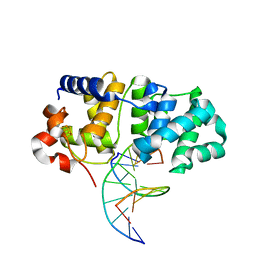 | | MutY N-terminal domain in complex with DNA containing an intrahelical oxoG:A base-pair | | Descriptor: | Adenine DNA glycosylase, CALCIUM ION, DNA (5'-D(*AP*GP*CP*AP*CP*AP*GP*GP*AP*T)-3'), ... | | Authors: | Wang, L, Chakravarthy, S, Verdine, G.L. | | Deposit date: | 2016-06-27 | | Release date: | 2017-02-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural Basis for the Lesion-scanning Mechanism of the MutY DNA Glycosylase.
J. Biol. Chem., 292, 2017
|
|
5KP0
 
 | | Recognition and targeting mechanisms by chaperones in flagella assembly and operation | | Descriptor: | Flagellar protein FliT,Flagellum-specific ATP synthase | | Authors: | Khanra, N.K, Rossi, P, Economou, A, Kalodimos, C.G. | | Deposit date: | 2016-07-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Recognition and targeting mechanisms by chaperones in flagellum assembly and operation.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5KSR
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa - XFSurE-TB (Tetramer Bigger). | | Descriptor: | 5'-nucleotidase SurE, CHLORIDE ION, IODIDE ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, De Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5L1H
 
 | | AMPA subtype ionotropic glutamate receptor GluA2 in complex with noncompetitive inhibitor GYKI53655 | | Descriptor: | (8R)-5-(4-aminophenyl)-N,8-dimethyl-8,9-dihydro-2H,7H-[1,3]dioxolo[4,5-h][2,3]benzodiazepine-7-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor 2 | | Authors: | Yelshanskaya, M.V, Singh, A.K, Sampson, J.M, Sobolevsky, A.I. | | Deposit date: | 2016-07-29 | | Release date: | 2016-10-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.801 Å) | | Cite: | Structural Bases of Noncompetitive Inhibition of AMPA-Subtype Ionotropic Glutamate Receptors by Antiepileptic Drugs.
Neuron, 91, 2016
|
|
5KK2
 
 | | Architecture of fully occupied GluA2 AMPA receptor - TARP complex elucidated by single particle cryo-electron microscopy | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Yoshioka, C, Baconguis, I, Gouaux, E. | | Deposit date: | 2016-06-20 | | Release date: | 2016-07-06 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of fully occupied GluA2 AMPA receptor-TARP complex elucidated by cryo-EM.
Nature, 536, 2016
|
|
5KJ4
 
 | |
5KN5
 
 | | TGFalpha/Epiregulin complex with neutralizing antibody LY3016859 | | Descriptor: | Epiregulin Antibody LY3016859 Fab Heavy Chain, Epiregulin Antibody LY3016859 Fab Light Chain, Protransforming growth factor alpha, ... | | Authors: | Atwell, S, Boyles, J.S, Clawson, D.K, Druzina, Z, Josef, G.H, Weichert, K, Witcher, D.R. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-31 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of selectivity and neutralizing activity of a TGF alpha /epiregulin specific antibody.
Protein Sci., 25, 2016
|
|
5KR7
 
 | | KDM4C bound to pyrazolo-pyrimidine scaffold | | Descriptor: | 6-ethyl-2,5-dimethyl-7-oxidanylidene-4~{H}-pyrazolo[1,5-a]pyrimidine-3-carbonitrile, CHLORIDE ION, FE (II) ION, ... | | Authors: | Bellon, S.F, Poy, F, Setser, J.W. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of potent, selective KDM5 inhibitors.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5KSQ
 
 | | Stationary phase survival protein E (SurE) from Xylella fastidiosa | | Descriptor: | 5'-nucleotidase SurE, IODIDE ION, MANGANESE (II) ION, ... | | Authors: | Machado, A.T.P, Fonseca, E.M.B, Dos Reis, M.A, Saraiva, A.M, Dos Santos, C.A, De Toledo, M.A, Polikarpov, I, De Souza, A.P, Aparicio, R, Iulek, J. | | Deposit date: | 2016-07-09 | | Release date: | 2017-07-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Conformational variability of the stationary phase survival protein E from Xylella fastidiosa revealed by X-ray crystallography, small-angle X-ray scattering studies, and normal mode analysis.
Proteins, 85, 2017
|
|
5L0F
 
 | |
5LAS
 
 | |
5L1P
 
 | | X-ray Structure of Cytochrome P450 PntM with Pentalenolactone | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Pentalenolactone synthase, pentalenolactone | | Authors: | Duan, L, Jogl, G, Cane, D.E. | | Deposit date: | 2016-07-29 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Cytochrome P450-Catalyzed Oxidative Rearrangement in the Final Step of Pentalenolactone Biosynthesis: Substrate Structure Determines Mechanism.
J.Am.Chem.Soc., 138, 2016
|
|
5KLM
 
 | | Crystal structure of 2-hydroxymuconate-6-semialdehyde derived intermediate in NAD(+)-bound 2-aminomuconate 6-semialdehyde dehydrogenase N169D | | Descriptor: | 2-aminomuconate 6-semialdehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION | | Authors: | Yang, Y, Davis, I, Ha, U, Wang, Y, Shin, I, Liu, A. | | Deposit date: | 2016-06-24 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | A Pitcher-and-Catcher Mechanism Drives Endogenous Substrate Isomerization by a Dehydrogenase in Kynurenine Metabolism.
J.Biol.Chem., 291, 2016
|
|
5KLF
 
 | | Structure of CBM_E1, a novel carbohydrate-binding module found by sugar cane soil metagenome, complexed with cellopentaose and gadolinium ion | | Descriptor: | Carbohydrate binding module E1, GADOLINIUM ATOM, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Liberato, M.V, Campos, B.M, Zeri, A.C.M, Squina, F.M. | | Deposit date: | 2016-06-24 | | Release date: | 2016-09-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | A Novel Carbohydrate-binding Module from Sugar Cane Soil Metagenome Featuring Unique Structural and Carbohydrate Affinity Properties.
J.Biol.Chem., 291, 2016
|
|
5KPF
 
 | | Crystal structure of cytochrome c - Phenyl-trisulfonatocalix[4]arene complex | | Descriptor: | Cytochrome c iso-1, HEME C, NITRATE ION, ... | | Authors: | Doolan, A.M, Rennie, M.L, Crowley, P.B. | | Deposit date: | 2016-07-04 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Protein Recognition by Functionalized Sulfonatocalix[4]arenes.
Chemistry, 24, 2018
|
|
5L5G
 
 | | Plexin A2 full extracellular region, domains 1 to 8 modeled, data to 10 angstrom | | Descriptor: | Plexin-A2 | | Authors: | Janssen, B.J.C, Kong, Y, Malinauskas, T, Vangoor, V.R, Coles, C.H, Kaufmann, R, Ni, T, Gilbert, R.J.C, Padilla-Parra, S, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2016-05-28 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (10 Å) | | Cite: | Structural Basis for Plexin Activation and Regulation.
Neuron, 91, 2016
|
|
5L8B
 
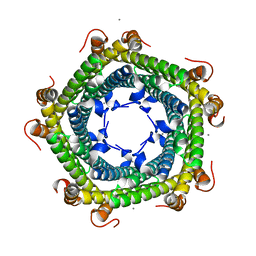 | | Crystal structure of Rhodospirillum rubrum Rru_A0973 mutant E62A | | Descriptor: | CALCIUM ION, Uncharacterized protein | | Authors: | He, D, Hughes, S, Vanden-Hehir, S, Georgiev, A, Altenbach, K, Tarrant, E, Mackay, C.L, Waldron, K.J, Clarke, D.J, Marles-Wright, J. | | Deposit date: | 2016-06-07 | | Release date: | 2016-08-31 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural characterization of encapsulated ferritin provides insight into iron storage in bacterial nanocompartments.
Elife, 5, 2016
|
|
