4QPP
 
 | | The Crystal Structure of Human HMT1 hnRNP methyltransferase-like protein 6 in complex with compound DS-421 (2-{4-[3-CHLORO-2-(2-METHOXYPHENYL)-1H-INDOL-5-YL]PIPERIDIN-1-YL}-N-METHYLETHANAMINE | | Descriptor: | 2-{4-[3-chloro-2-(2-methoxyphenyl)-1H-indol-5-yl]piperidin-1-yl}-N-methylethanamine, POLY-UNK, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, Smil, D, Walker, J.R, He, H, Eram, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-24 | | Release date: | 2014-08-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Crystal Structure of Human HMT1
hnRNP methyltransferase-like protein 6 in complex with compound DS-421
To be Published
|
|
6Y09
 
 | |
8P1V
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 6-cyclopropyl-~{N}-(2-methylindazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
5K2L
 
 | | Crystal structure of LysM domain from Volvox carteri chitinase | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, lysozyme | | Authors: | Kitaoku, Y, Numata, T, Fukamizo, T, Ohnuma, T. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Chitin oligosaccharide binding to the lysin motif of a novel type of chitinase from the multicellular green alga, Volvox carteri.
Plant Mol. Biol., 93, 2017
|
|
8P1W
 
 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with allosteric compound STL232591 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, 8-methoxy-1-(4-methoxyphenyl)-3-methyl-2-oxidanyl-[1]benzofuro[3,2-c]pyridine, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
7P5P
 
 | | Pyrazole Carboxylic Acid Inhibitors of KEAP1:NRF2 interaction | | Descriptor: | 5-[(1~{R},2~{R})-2-(1-methyl-1,2,3-triazol-4-yl)cyclopropyl]-1-[3-[3-[(2~{R})-2-propylpiperidin-1-yl]carbonylphenyl]phenyl]pyrazole-4-carboxylic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G. | | Deposit date: | 2021-07-14 | | Release date: | 2021-11-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Guided Discovery of Pyrazole Carboxylic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1: Nuclear Factor Erythroid 2 Related Factor 2 (KEAP1:NRF2) Protein-Protein Interaction.
J.Med.Chem., 64, 2021
|
|
8RQD
 
 | | Crystal structure of human DNPH1 mutant-Y24F | | Descriptor: | 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Devi, S, da Silva, R.G. | | Deposit date: | 2024-01-17 | | Release date: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Human 2'-Deoxynucleoside 5'-Phosphate N-Hydrolase 1: The Catalytic Roles of Tyr24 and Asp80.
Chembiochem, 25, 2024
|
|
7ZU3
 
 | |
7MBH
 
 | | Structure of Human Enolase 2 in complex with phosphoserine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Gamma-enolase, ... | | Authors: | Leonard, P.G, Hicks, K.G, Rutter, J. | | Deposit date: | 2021-03-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Protein-metabolite interactomics of carbohydrate metabolism reveal regulation of lactate dehydrogenase.
Science, 379, 2023
|
|
6W5L
 
 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | Descriptor: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
4REE
 
 | | Crystal Structure of TR3 LBD in complex with Molecule 6 | | Descriptor: | 1-(2,3,4-trihydroxyphenyl)nonan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Wang, W.J, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
8F6C
 
 | | E. coli cytochrome bo3 ubiquinol oxidase dimer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
8F68
 
 | | E. coli cytochrome bo3 ubiquinol oxidase monomer | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, COPPER (II) ION, Cytochrome bo(3) ubiquinol oxidase subunit 1, ... | | Authors: | Guo, Y, Karimullina, E, Borek, D, Savchenko, A, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2022-11-16 | | Release date: | 2022-11-30 | | Last modified: | 2025-09-17 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Monomer and dimer structures of cytochrome bo 3 ubiquinol oxidase from Escherichia coli.
Protein Sci., 32, 2023
|
|
6O98
 
 | | Crystal structure of RAR-related orphan receptor C in complex with a phenyl (3-phenylpyrrolidin-3-yl)sulfone inhibitor | | Descriptor: | 1-(4-{(3R)-3-[(4-fluorophenyl)sulfonyl]-3-[4-(1,1,1,3,3,3-hexafluoro-2-hydroxypropan-2-yl)phenyl]pyrrolidine-1-carbonyl}piperazin-1-yl)ethan-1-one, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2019-03-13 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-based Discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as Selective, Orally Active ROR gamma t Inverse Agonists.
ACS Med Chem Lett, 10, 2019
|
|
8I5S
 
 | | Crystal structure of TxGH116 D593N acid/base mutant from Thermoanaerobacterium xylanolyticum with 2-deoxy-2-fluoroglucoside | | Descriptor: | 1,2-ETHANEDIOL, 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, ... | | Authors: | Pengthaisong, S, Ketudat Cairns, J.R. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reaction Mechanism of Glycoside Hydrolase Family 116 Utilizes Perpendicular Protonation.
Acs Catalysis, 13, 2023
|
|
6TT5
 
 | | Crystal structure of DCLRE1C/Artemis | | Descriptor: | 1,2-ETHANEDIOL, NICKEL (II) ION, Protein artemis, ... | | Authors: | Yosaatmadja, Y, Goubin, S, Newman, J.A, Mukhopadhyay, S.M.M, Dannerfjord, A.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-12-23 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic insights into the Artemis endonuclease and strategies for its inhibition.
Nucleic Acids Res., 49, 2021
|
|
8I5R
 
 | |
7MCM
 
 | |
8G8D
 
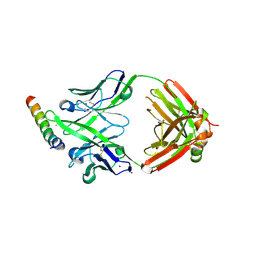 | | Crystal structure of DH1346 Fab in complex with HIV proximal MPER peptide | | Descriptor: | DH1346 heavy chain, DH1346 light chain, FLUORIDE ION, ... | | Authors: | Niyongabo, A, Janus, B.M, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
8G8A
 
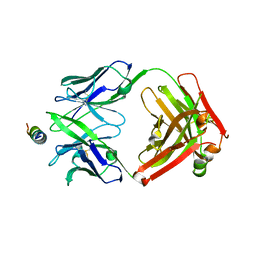 | | Crystal structure of DH1317.8 Fab in complex with HIV proximal MPER peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH1317.8 heavy chain, DH1317.8 light chain, ... | | Authors: | Janus, B.M, Astavans, A, Ofek, G. | | Deposit date: | 2023-02-17 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Vaccine induction of heterologous HIV-1-neutralizing antibody B cell lineages in humans.
Cell, 187, 2024
|
|
6YGB
 
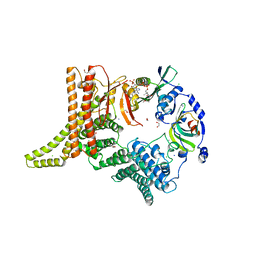 | | Crystal structure of the NatC complex bound to CoA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, COENZYME A, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
6VW9
 
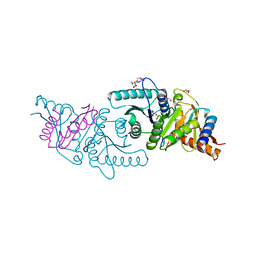 | | C-terminal regulatory domain of the chloride transporter KCC-1 from C. elegans, proteolyzed during crystallization | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, K+/Cl-Cotransporter | | Authors: | Zimanyi, C.M, Cheung, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Regulatory Cytosolic Domain of a Eukaryotic Potassium-Chloride Cotransporter.
Structure, 28, 2020
|
|
8G3C
 
 | |
7ZQ7
 
 | | Structure of RpF-1 | | Descriptor: | Crotonase/enoyl-CoA hydratase family protein | | Authors: | Sanchez-Alba, L, Reverter, D, Conchillo, O, Yero, D, Daura, X, Gibert, I. | | Deposit date: | 2022-04-29 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of RpF-1
To Be Published
|
|
8D1I
 
 | | hBest1 1uM Ca2+ (Ca2+-bound) closed state | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Bestrophin-1, CALCIUM ION | | Authors: | Owji, A.P, Kittredge, A, Hendrickson, W.A, Tingting, Y. | | Deposit date: | 2022-05-27 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.82 Å) | | Cite: | Structures and gating mechanisms of human bestrophin anion channels.
Nat Commun, 13, 2022
|
|
