8C27
 
 | |
8AUP
 
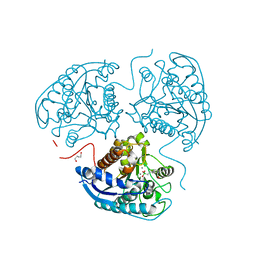 | | Structure of hARG1 with a novel inhibitor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[(1~{R},3~{R},4~{S})-3-azanyl-3-carboxy-4-[(dimethylamino)methyl]cyclohexyl]ethyl-$l^{3}-oxidanyl-bis(oxidanyl)boron, Arginase-1, ... | | Authors: | Napiorkowska-Gromadzka, A, Nowak, E, Nowotny, M. | | Deposit date: | 2022-08-25 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Arginase 1/2 Inhibitor OATD-02: From Discovery to First-in-man Setup in Cancer Immunotherapy.
Mol.Cancer Ther., 22, 2023
|
|
8TXA
 
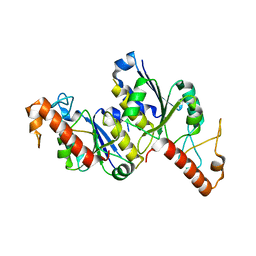 | | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Balsamo, A, Bruno, C, Edele, C, Fabian, T, Jannotta, R, Lee, R, Schryver, D, Warsaw, J, Warsaw, L, Stojanoff, V, Battaile, K, Perez, A, Bolen, R. | | Deposit date: | 2023-08-23 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.591 Å) | | Cite: | Apo Structure of (N1G37) tRNA Methyltransferase from Mycobacterium marinum
To Be Published
|
|
6XS6
 
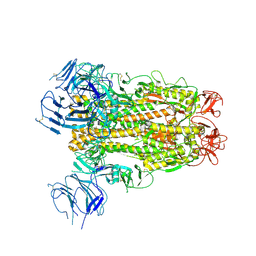 | | SARS-CoV-2 Spike D614G variant, minus RBD | | Descriptor: | Spike glycoprotein | | Authors: | Wang, X, Egri, S.B, Dudkina, N, Luban, J, Shen, K. | | Deposit date: | 2020-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant.
Cell, 183, 2020
|
|
6JW7
 
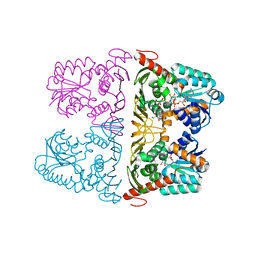 | | The crystal structure of KanD2 in complex with NADH and 3"-deamino-3"-hydroxykanamycin A | | Descriptor: | (2R,3S,4S,5R,6R)-2-(aminomethyl)-6-[(1R,2S,3S,4R,6S)-4,6-bis(azanyl)-3-[(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-2-oxidanyl-cyclohexyl]oxy-oxane-3,4,5-triol, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Dehydrogenase | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
6JW6
 
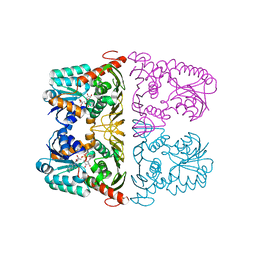 | | The crystal structure of KanD2 in complex with NAD | | Descriptor: | Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kudo, F, Kitayama, Y, Miyanaga, A, Hirayama, A, Eguchi, T. | | Deposit date: | 2019-04-18 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Biochemical and structural analysis of a dehydrogenase, KanD2, and an aminotransferase, KanS2, that are responsible for the construction of the kanosamine moiety in kanamycin biosynthesis.
Biochemistry, 59, 2020
|
|
5VEQ
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Y | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VER
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2Z | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5VEP
 
 | | MOUSE KYNURENINE AMINOTRANSFERASE III, RE-REFINEMENT OF THE PDB STRUCTURE 3E2F | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wlodawer, A, Dauter, Z, Minor, W, Stanfield, R, Porebski, P, Jaskolski, M, Pozharski, E, Weichenberger, C.X, Rupp, B. | | Deposit date: | 2017-04-05 | | Release date: | 2017-11-29 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Detect, correct, retract: How to manage incorrect structural models.
FEBS J., 285, 2018
|
|
5O98
 
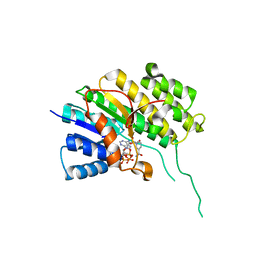 | | Binary complex of Catharanthus roseus Vitrosamine Synthase with NADP+ | | Descriptor: | Alcohol dehydrogenase 1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Stavrinides, A.K, Tatsis, E.C, Dang, T.T, Caputi, L, Stevenson, C.E.M, Lawson, D.M, Schneider, B, O'Connor, S.E. | | Deposit date: | 2017-06-16 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of a Short-Chain Dehydrogenase from Catharanthus roseus that Produces a New Monoterpene Indole Alkaloid.
Chembiochem, 19, 2018
|
|
5M5E
 
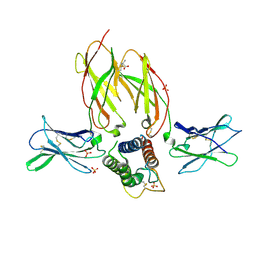 | | Crystal structure of a interleukin-2 variant in complex with interleukin-2 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klein, C, Freimoser-Grundschober, A, Waldhauer, I, Stihle, M, Birk, M, Benz, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines.
Oncoimmunology, 6, 2017
|
|
6MSU
 
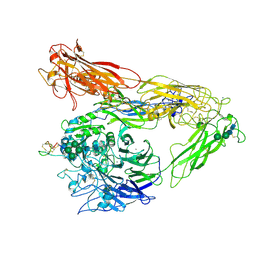 | | Integrin alphaVBeta3 in complex with EETI-II 2.5F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | van Agthoven, J.F, Arnaout, M.A. | | Deposit date: | 2018-10-18 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structural Basis of the Differential Binding of Engineered Knottins to Integrins alpha V beta 3 and alpha 5 beta 1.
Structure, 27, 2019
|
|
6CZ2
 
 | |
1CGX
 
 | |
3D4V
 
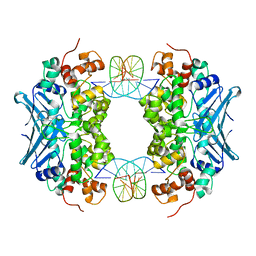 | | Crystal Structure of an AlkA Host/Guest Complex N7MethylGuanine:Cytosine Base Pair | | Descriptor: | 5'-D(*DGP*DAP*DCP*DAP*DTP*DGP*DAP*(FMG)P*DTP*DGP*DCP*DC)-3', 5'-D(*DGP*DGP*DCP*DAP*DCP*DTP*DCP*DAP*DTP*DGP*DTP*DC)-3', DNA-3-methyladenine glycosylase 2 | | Authors: | Lee, S, Bowman, B.R, Wang, S, Verdine, G.L. | | Deposit date: | 2008-05-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synthesis and structure of duplex DNA containing the genotoxic nucleobase lesion N7-methylguanine.
J.Am.Chem.Soc., 130, 2008
|
|
1CGW
 
 | |
1CGV
 
 | |
7RL3
 
 | | Structure of Drosophila melanogaster Plk4 PB3 | | Descriptor: | Serine/threonine-protein kinase PLK4 | | Authors: | Slep, K.C, Slevin, L.K. | | Deposit date: | 2021-07-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Polo-like kinase 4 homodimerization and condensate formation regulate its own protein levels but are not required for centriole assembly.
Mol.Biol.Cell, 34, 2023
|
|
8BUZ
 
 | | Structure of Adenylyl cyclase 8 bound to stimulatory G-protein, Ca2+/Calmodulin, Forskolin and MANT-GTP | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 8, FORSKOLIN, ... | | Authors: | Khanppnavar, B, Korkhov, V.M, Mehta, V. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.
Embo Rep., 25, 2024
|
|
8BV5
 
 | | Focus refinement of soluble domain of Adenylyl cyclase 8 bound to stimulatory G protein, Forskolin, ATPalphaS, and Ca2+/Calmodulin in lipid nanodisc conditions | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase type 8, FORSKOLIN, ... | | Authors: | Khanppnavar, B, Korkhov, V.M. | | Deposit date: | 2023-01-04 | | Release date: | 2024-01-17 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Regulatory sites of CaM-sensitive adenylyl cyclase AC8 revealed by cryo-EM and structural proteomics.
Embo Rep., 25, 2024
|
|
7KIX
 
 | |
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAW
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the complete 55S ribosome. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 16S ribosomal RNA, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-22 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
6GAZ
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 28S ribosomal subunit. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 28S ribosomal protein S18b, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
4CVQ
 
 | | CRYSTAL STRUCTURE OF AN AMINOTRANSFERASE FROM ESCHERICHIA COLI AT 2. 11 ANGSTROEM RESOLUTION | | Descriptor: | ACETATE ION, GLUTAMATE-PYRUVATE AMINOTRANSFERASE ALAA, GLYCEROL, ... | | Authors: | Penya-Soler, E, Fernandez, F.J, Lopez-Estepa, M, Garces, F, Richardson, A.J, Rudd, K.E, Coll, M, Vega, M.C. | | Deposit date: | 2014-03-28 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural analysis and mutant growth properties reveal distinctive enzymatic and cellular roles for the three major L-alanine transaminases of Escherichia coli.
PLoS ONE, 9, 2014
|
|
