7P16
 
 | |
7OJI
 
 | | ABCG2 topotecan turnover-2 state | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, ... | | Authors: | Yu, Q, Ni, D, Kowal, J, Manolaridis, I, Jackson, S.M, Stahlberg, H, Locher, K.P. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of ABCG2 under turnover conditions reveal a key step in the drug transport mechanism.
Nat Commun, 12, 2021
|
|
7P14
 
 | | Structure of full-length rXKR9 in complex with a sybody at 3.66A | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, Sybody, ... | | Authors: | Straub, M.S, Sawicka, M, Dutzler, R. | | Deposit date: | 2021-07-01 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Cryo-EM structures of the caspase activated protein XKR9 involved in apoptotic lipid scrambling.
Elife, 10, 2021
|
|
7P0Z
 
 | | 2.43 A Mycobacterium marinum EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Ye, G, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
7OM8
 
 | |
7OUG
 
 | | STLV-1 intasome:B56 in complex with the strand-transfer inhibitor raltegravir | | Descriptor: | DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), Integrase, ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OUH
 
 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor bictegravir | | Descriptor: | Bictegravir, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7P13
 
 | | 2.29 A Mycobacterium tuberculosis EspB. | | Descriptor: | ESX-1 secretion-associated protein EspB | | Authors: | Gijsbers, A, Zhang, Y, Vinciauskaite, V, Siroy, A, Gao, Y, Tria, G, Mathew, A, Sanchez-Puig, N, Lopez-Iglesias, C, Peters, P.J, Ravelli, R.B.G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Priming mycobacterial ESX-secreted protein B to form a channel-like structure.
Curr Res Struct Biol, 3, 2021
|
|
7OUF
 
 | | Structure of the STLV intasome:B56 complex bound to the strand-transfer inhibitor XZ450 | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[3-(dimethylamino)-3-oxidanylidene-propyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*CP*TP*GP*TP*GP*TP*TP*TP*GP*GP*CP*GP*CP*TP*TP*CP*TP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*AP*GP*AP*AP*GP*CP*GP*CP*CP*AP*AP*AP*CP*AP*CP*A)-3'), ... | | Authors: | Barski, M.S, Ballandras-Colas, A, Cronin, N.B, Pye, V.E, Cherepanov, P, Maertens, G.N. | | Deposit date: | 2021-06-11 | | Release date: | 2021-08-18 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the inhibition of HTLV-1 integration inferred from cryo-EM deltaretroviral intasome structures.
Nat Commun, 12, 2021
|
|
7OK0
 
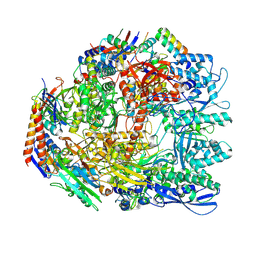 | | Cryo-EM structure of the Sulfolobus acidocaldarius RNA polymerase at 2.88 A | | Descriptor: | Conserved protein, DNA-directed RNA polymerase subunit A', DNA-directed RNA polymerase subunit A'', ... | | Authors: | Pilotto, S, Fouqueau, T, Lukoyanova, N, Sheppard, C, Lucas-Staat, S, Diaz-Santin, L.M, Matelska, D, Prangishvili, D, Cheung, A.C.M, Werner, F. | | Deposit date: | 2021-05-17 | | Release date: | 2021-08-25 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of RNA polymerase inhibition by viral and host factors.
Nat Commun, 12, 2021
|
|
7OYG
 
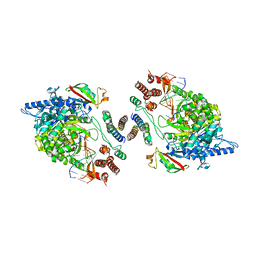 | | Dimeric form of SARS-CoV-2 RNA-dependent RNA polymerase | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase (nsp12), ... | | Authors: | Jochheim, F.A, Tegunov, D, Hillen, H.S, Schmitzova, J, Kokic, G, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | The structure of a dimeric form of SARS-CoV-2 polymerase
Communications Biology, 4, 2021
|
|
8V5M
 
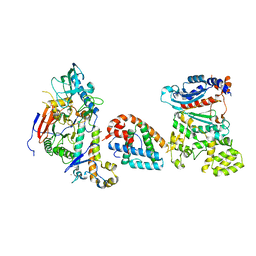 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6J
 
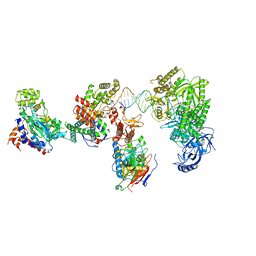 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5O
 
 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6I
 
 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6G
 
 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
7OTQ
 
 | | Cryo-EM structure of ALC1/CHD1L bound to a PARylated nucleosome | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (149-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2021-06-10 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and dynamics of the chromatin remodeler ALC1 bound to a PARylated nucleosome
Elife, 10, 2021
|
|
8UO1
 
 | | CryoEM structure of beta-2-adrenergic receptor in complex with GTP-bound Gs heterotrimer (Class Q) | | Descriptor: | (5R,6R)-6-(methylamino)-5,6,7,8-tetrahydronaphthalene-1,2,5-triol, Beta-2 adrenergic receptor, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Papasergi-Scott, M.M, Skiniotis, G. | | Deposit date: | 2023-10-19 | | Release date: | 2024-03-06 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Time-resolved cryo-EM of G-protein activation by a GPCR.
Nature, 629, 2024
|
|
7OSE
 
 | | cytochrome bd-II type oxidase with bound aurachin D | | Descriptor: | Aurachin D, CIS-HEME D HYDROXYCHLORIN GAMMA-SPIROLACTONE, Cytochrome bd-II ubiquinol oxidase subunit 1, ... | | Authors: | Grauel, A, Kaegi, J, Rasmussen, T, Wohlwend, D, Boettcher, B, Friedrich, T. | | Deposit date: | 2021-06-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of Escherichia coli cytochrome bd-II type oxidase with bound aurachin D.
Nat Commun, 12, 2021
|
|
7ODS
 
 | | State B of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, DNA (30-MER),16S mitochondrial rRNA, 39S ribosomal protein L10, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
7ODT
 
 | | State C of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
7ODR
 
 | | State A of the human mitoribosomal large subunit assembly intermediate | | Descriptor: | 16S mitochondrial rRNA, DNA (31-MER),16S mitochondrial rRNA, 39S ribosomal protein L10, ... | | Authors: | Lenarcic, T, Jaskolowski, M, Leibundgut, M, Scaiola, A, Schoenhut, T, Saurer, M, Lee, R.G, Rackham, O, Filipovska, A, Ban, N. | | Deposit date: | 2021-04-30 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Stepwise maturation of the peptidyl transferase region of human mitoribosomes.
Nat Commun, 12, 2021
|
|
7NWI
 
 | | Mammalian pre-termination 80S ribosome with Empty-A site bound by Blasticidin S | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA+BlaS, 40S ribosomal protein S11, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
