7KJ7
 
 | | Plasmodium falciparum protein Pf12p | | Descriptor: | Surface protein P12p | | Authors: | Dietrich, M.H, Tham, W.H. | | Deposit date: | 2020-10-26 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Nanobody generation and structural characterization of Plasmodium falciparum 6-cysteine protein Pf12p.
Biochem.J., 478, 2021
|
|
5AWH
 
 | | Rhodobacter sphaeroides Argonaute in complex with guide RNA/target DNA heteroduplex | | Descriptor: | DNA (5'-D(*CP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*A)-3'), MAGNESIUM ION, RNA (5'-D(P*UP*UP*AP*CP*AP*AP*CP*CP*UP*AP*CP*UP*AP*CP*CP*UP*CP*G)-3'), ... | | Authors: | Miyoshi, T, Ito, K, Murakami, R, Uchiumi, T. | | Deposit date: | 2015-07-03 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute.
Nat Commun, 7, 2016
|
|
5O9A
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J-L504Y-N775S) in complex with glutamate and BPAM121 at 1.78 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 7-chloro-4-(2-fluoroethyl)-2,3-dihydro-1,2,4-benzothiadiazine 1,1-dioxide, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Rovinskaja, K, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-06-16 | | Release date: | 2018-01-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
7KOE
 
 | | Electron bifurcating flavoprotein Fix/EtfABCX | | Descriptor: | Electron transfer flavoprotein, alpha subunit, beta subunit, ... | | Authors: | Feng, X, Li, H. | | Deposit date: | 2020-11-08 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryoelectron microscopy structure and mechanism of the membrane-associated electron-bifurcating flavoprotein Fix/EtfABCX.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5OBR
 
 | | Aurora A kinase in complex with 2-(3-chloro-5-fluorophenyl)quinoline-4-carboxylic acid and JNJ-7706621 | | Descriptor: | 2-(3-chloranyl-5-fluoranyl-phenyl)quinoline-4-carboxylic acid, 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
5UZB
 
 | | Cryo-EM structure of the MAL TIR domain filament | | Descriptor: | Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Ve, T, Vajjhala, P.R, Hedger, A, Croll, T, DiMaio, F, Horsefield, S, Yu, X, Lavrencic, P, Hassan, Z, Morgan, G.P, Mansell, A, Mobli, M, O'Carrol, A, Chauvin, B, Gambin, Y, Sierecki, E, Landsberg, M.J, Stacey, K.J, Egelman, E.H, Kobe, B. | | Deposit date: | 2017-02-25 | | Release date: | 2017-07-26 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Structural basis of TIR-domain-assembly formation in MAL- and MyD88-dependent TLR4 signaling.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5OCS
 
 | | Ene-reductase (ER/OYE) from Ralstonia (Cupriavidus) metallidurans | | Descriptor: | ACETATE ION, CITRIC ACID, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Opperman, D.J. | | Deposit date: | 2017-07-03 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural investigation into the C-terminal extension of the ene-reductase from Ralstonia (Cupriavidus) metallidurans.
Proteins, 85, 2017
|
|
5OEW
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with glutamate and positive allosteric modulator BPAM538 | | Descriptor: | 4-cyclopropyl-7-(3-methoxyphenoxy)-2,3-dihydro-1$l^{6},2,4-benzothiadiazine 1,1-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Frydenvang, K.A, Kastrup, J.S. | | Deposit date: | 2017-07-10 | | Release date: | 2018-01-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 7-Phenoxy-Substituted 3,4-Dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides as Positive Allosteric Modulators of alpha-Amino-3-hydroxy-5-methyl-4-isoxazolepropionic Acid (AMPA) Receptors with Nanomolar Potency.
J. Med. Chem., 61, 2018
|
|
5A8B
 
 | | Structure of a parallel dimer of the aureochrome 1a LOV domain from Phaeodactylum tricornutum | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Banerjee, A, Herman, E, Kottke, T, Essen, L.O. | | Deposit date: | 2015-07-14 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.791 Å) | | Cite: | Structure of a Native-Like Aureochrome 1A Lov Domain Dimer from Phaeodactylum Tricornutum.
Structure, 24, 2016
|
|
5A8F
 
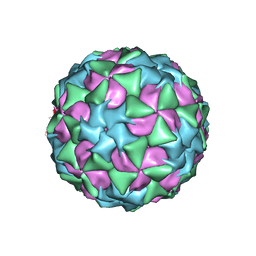 | | Structure and genome release mechanism of human cardiovirus Saffold virus-3 | | Descriptor: | GENOME POLYPHUMAN SAFFOLD VIRUS-3 VP3 PROTEIN, HUMAN SAFFOLD VIRUS-3 VP1, HUMAN SAFFOLD VIRUS-3 VP2 | | Authors: | Mullapudi, E, Novacek, J, Palkova, L, Kulich, P, Lindberg, M, vanKuppeveld, F.J.M, Plevka, P. | | Deposit date: | 2015-07-15 | | Release date: | 2016-06-08 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (10.6 Å) | | Cite: | Structure and Genome Release Mechanism of Human Cardiovirus Saffold Virus-3.
J.Virol., 90, 2016
|
|
7UN6
 
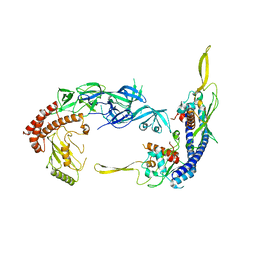 | | Complex of UBE2O with NAP1L1 | | Descriptor: | (E3-independent) E2 ubiquitin-conjugating enzyme, Nucleosome assembly protein 1-like 1 | | Authors: | Yip, M.C.J, Sedor, S.F, Shao, S. | | Deposit date: | 2022-04-09 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of client selection by the protein quality-control factor UBE2O.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5OID
 
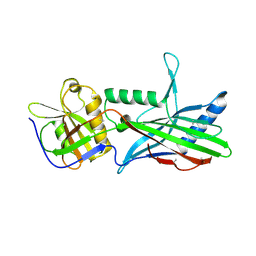 | | Complex Trichoplax STIL-NTD:human CEP85 coiled coil domain 4 | | Descriptor: | Centrosomal protein of 85 kDa, Putative uncharacterized protein | | Authors: | van Breugel, M. | | Deposit date: | 2017-07-18 | | Release date: | 2018-04-18 | | Last modified: | 2018-05-09 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Direct binding of CEP85 to STIL ensures robust PLK4 activation and efficient centriole assembly.
Nat Commun, 9, 2018
|
|
5OKM
 
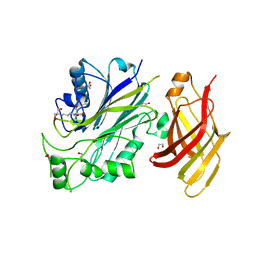 | | Crystal structure of human SHIP2 Phosphatase-C2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NITRATE ION, ... | | Authors: | Le Coq, J, Lietha, D. | | Deposit date: | 2017-07-25 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural basis for interdomain communication in SHIP2 providing high phosphatase activity.
Elife, 6, 2017
|
|
5UQM
 
 | |
4ZZI
 
 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
7FVS
 
 | | Crystal Structure of S. aureus gyrase in complex with 4-[[1-[(1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl)methyl]azetidin-3-yl]methylamino]-6-fluorochromen-2-one | | Descriptor: | 4-{[(1-{[(6R)-1-chloro-6,7-dihydro-5H-cyclopenta[c]pyridin-6-yl]methyl}azetidin-3-yl)methyl]amino}-6-fluoro-2H-1-benzopyran-2-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Xu, B, Benz, J, Cumming, J.G, Kreis, L, Rudolph, M.G. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
5OLM
 
 | | TRIM21 | | Descriptor: | E3 ubiquitin-protein ligase TRIM21, ZINC ION | | Authors: | James, L.C. | | Deposit date: | 2017-07-28 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Intracellular antibody signalling is regulated by phosphorylation of the Fc receptor TRIM21.
Elife, 7, 2018
|
|
8AFV
 
 | | DaArgC3 - Engineered Formyl Phosphate Reductase with 3 substitutions (S178V, G182V, L233I) | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
7FVT
 
 | | Crystal Structure of S. aureus gyrase in complex with 6-[5-[2-[(4-chloro-2,3-dihydro-1H-inden-2-yl)methylamino]ethyl]-2-oxo-1,3-oxazolidin-3-yl]-4H-pyrido[3,2-b][1,4]oxazin-3-one | | Descriptor: | 6-{(5R)-5-[2-({[(2R)-4-chloro-2,3-dihydro-1H-inden-2-yl]methyl}amino)ethyl]-2-oxo-1,3-oxazolidin-3-yl}-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, CHLORIDE ION, DNA (5'-D(*AP*GP*CP*CP*GP*TP*AP*GP*GP*GP*CP*CP*CP*TP*AP*CP*GP*GP*CP*T)-3'), ... | | Authors: | Xu, B, Benz, J, Cumming, J.G, Rudolph, M.G. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.081 Å) | | Cite: | Discovery of a Series of Indane-Containing NBTIs with Activity against Multidrug-Resistant Gram-Negative Pathogens.
Acs Med.Chem.Lett., 14, 2023
|
|
7UN3
 
 | | Complex of UBE2O with NAP1L1 and ubiquitylated uL2 | | Descriptor: | Nucleosome assembly protein 1-like 1, Ubiquitin,60S ribosomal protein L8,(E3-independent) E2 ubiquitin-conjugating enzyme fusion | | Authors: | Yip, M.C.J, Sedor, S.F, Shao, S. | | Deposit date: | 2022-04-08 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mechanism of client selection by the protein quality-control factor UBE2O.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8AFU
 
 | | DaArgC - N-acetyl-gamma-glutamyl-phosphate Reductase of Denitrovibrio acetiphilus | | Descriptor: | N-acetyl-gamma-glutamyl-phosphate reductase, SODIUM ION | | Authors: | Pfister, P, Nattermann, M, Zarzycki, J, Erb, T.J. | | Deposit date: | 2022-07-18 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Engineering a new-to-nature cascade for phosphate-dependent formate to formaldehyde conversion in vitro and in vivo.
Nat Commun, 14, 2023
|
|
8AVE
 
 | | Human leptin in complex with the human LEP-R ectodomain fused to a C-terminal trimeric isoleucine GCN4 zipper (2:2 model) | | Descriptor: | Leptin, Leptin receptor | | Authors: | Verstraete, K, Savvides, S.N, Verschueren, K.G, Tsirigotaki, A. | | Deposit date: | 2022-08-26 | | Release date: | 2023-04-05 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (5.62 Å) | | Cite: | Mechanism of receptor assembly via the pleiotropic adipokine Leptin.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5VYZ
 
 | | Crystal structure of Lactococcus lactis pyruvate carboxylase in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8AG8
 
 | |
5O85
 
 | | p34-p44 complex | | Descriptor: | General transcription factor IIH subunit 2, General transcription factor IIH subunit 3, ZINC ION | | Authors: | Radu, L, Poterszman, A. | | Deposit date: | 2017-06-12 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
