1IGM
 
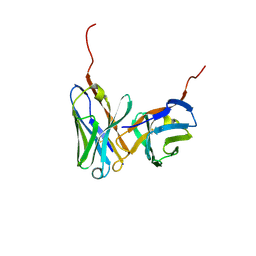 | |
3UMK
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with copper | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
3UWB
 
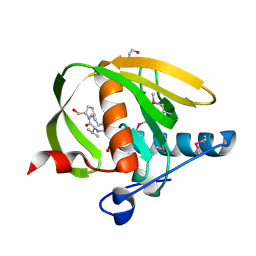 | | Crystal structure of a probable peptide deformylase from strucynechococcus phage S-SSM7 in complex with actinonin | | Descriptor: | 1,2-ETHANEDIOL, ACTINONIN, CHLORIDE ION, ... | | Authors: | Lorimer, D, Abendroth, J, Edwards, T.E, Burgin, A, Segall, A, Rohwer, F. | | Deposit date: | 2011-12-01 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and function of a cyanophage-encoded peptide deformylase.
ISME J, 7, 2013
|
|
7ZQX
 
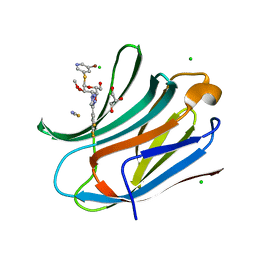 | | Human galectin-3c in complex with a galactose derivative | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{R})-2-(5-bromanylpyridin-3-yl)sulfanyl-6-(hydroxymethyl)-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zetterberg, F, Nilsson, U.J, Hakansson, M, Logan, D.T. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Discovery and Optimization of the First Highly Effective and Orally Available Galectin-3 Inhibitors for Treatment of Fibrotic Disease.
J.Med.Chem., 65, 2022
|
|
3UWL
 
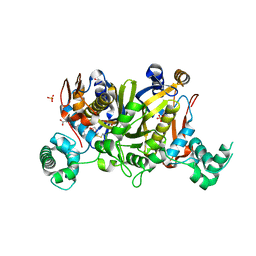 | | Crystal structure of Enteroccocus faecalis thymidylate synthase (EfTS) in complex with 5-formyl tetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SULFATE ION, ... | | Authors: | Pozzi, C, Catalano, A, Cortesi, D, Luciani, R, Ferrari, S, Fritz, T, Costi, M.P, Mangani, S. | | Deposit date: | 2011-12-02 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The structure of Enterococcus faecalis thymidylate synthase provides clues about folate bacterial metabolism.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1IZN
 
 | |
1IH5
 
 | | CRYSTAL STRUCTURE OF AQUAPORIN-1 | | Descriptor: | AQUAPORIN-1 | | Authors: | Ren, G, Reddy, V.S, Cheng, A, Melnyk, P, Mitra, A.K. | | Deposit date: | 2001-04-18 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | ELECTRON CRYSTALLOGRAPHY (3.7 Å) | | Cite: | Visualization of a water-selective pore by electron crystallography in vitreous ice.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
3V1Y
 
 | | Crystal structures of glyceraldehyde-3-phosphate dehydrogenase complexes with NAD | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, cytosolic, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Tien, Y.C, Chuankhayan, P, Lin, Y.H, Chang, S.L, Chen, C.J. | | Deposit date: | 2011-12-10 | | Release date: | 2012-11-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structures of rice (Oryza sativa) glyceraldehyde-3-phosphate dehydrogenase complexes with NAD and sulfate suggest involvement of Phe37 in NAD binding for catalysis
Plant Mol.Biol., 80, 2012
|
|
1II5
 
 | |
4X3H
 
 | | CRYSTAL STRUCTURE OF ARC N-LOBE COMPLEXED WITH STARGAZIN PEPTIDE | | Descriptor: | Activity-regulated cytoskeleton-associated protein, VOLTAGE-DEPENDENT CALCIUM CHANNEL GAMMA-2 SUBUNIT | | Authors: | zhang, W, ward, m, leahy, d, worley, p. | | Deposit date: | 2014-11-30 | | Release date: | 2015-06-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural basis of arc binding to synaptic proteins: implications for cognitive disease.
Neuron, 86, 2015
|
|
1J6Y
 
 | | Solution structure of Pin1At from Arabidopsis thaliana | | Descriptor: | peptidyl-prolyl cis-trans isomerase | | Authors: | Landrieu, I, Wieruszeski, J.M, Wintjens, R, Inze, D, Lippens, G. | | Deposit date: | 2001-05-15 | | Release date: | 2002-08-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the
Single-domain Prolyl Cis/Trans
Isomerase PIN1At from Arabidopsis thaliana
J.Mol.Biol., 320, 2002
|
|
3V5W
 
 | |
1IH9
 
 | | NMR Structure of Zervamicin IIB (peptaibol antibiotic) Bound to DPC Micelles | | Descriptor: | ZERVAMICIN IIB | | Authors: | Shenkarev, Z.O, Balasheva, T.A, Efremov, R.G, Yakimenko, Z.A, Ovchinnikova, T.V, Raap, J, Arseniev, A.S. | | Deposit date: | 2001-04-19 | | Release date: | 2002-02-13 | | Last modified: | 2012-12-12 | | Method: | SOLUTION NMR | | Cite: | Spatial Structure of Zervamicin Iib Bound to Dpc Micelles: Implications for Voltage-Gating.
Biophys.J., 82, 2002
|
|
3UH2
 
 | | Tankyrase-1 in complexed with PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, SULFATE ION, Tankyrase-1, ... | | Authors: | Kirby, C.A, Stams, T. | | Deposit date: | 2011-11-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of human tankyrase 1 in complex with small-molecule inhibitors PJ34 and XAV939.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
1IO5
 
 | | HYDROGEN AND HYDRATION OF HEN EGG-WHITE LYSOZYME DETERMINED BY NEUTRON DIFFRACTION | | Descriptor: | LYSOZYME C | | Authors: | Niimura, N, Minezaki, Y, Nonaka, T, Castagna, J.C, Cipriani, F, Hoeghoej, P, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 2001-01-14 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Neutron Laue diffractometry with an imaging plate provides an effective data collection regime for neutron protein crystallography.
Nat.Struct.Biol., 4, 1997
|
|
3UPY
 
 | | Crystal structure of the Brucella abortus enzyme catalyzing the first committed step of the methylerythritol 4-phosphate pathway. | | Descriptor: | 3-[FORMYL(HYDROXY)AMINO]PROPYLPHOSPHONIC ACID, MAGNESIUM ION, Oxidoreductase | | Authors: | Calisto, B.M, Perez-Gil, J, Fita, I, Rodriguez-Concepcion, M. | | Deposit date: | 2011-11-18 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Brucella abortus deoxyxylulose-5-phosphate reductoisomerase-like (DRL) enzyme involved in isoprenoid biosynthesis.
J.Biol.Chem., 287, 2012
|
|
3USY
 
 | |
4XP4
 
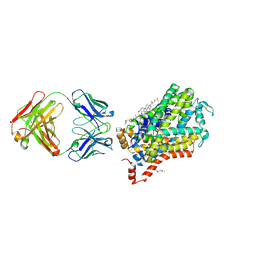 | | X-ray structure of Drosophila dopamine transporter in complex with cocaine | | Descriptor: | 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ANTIBODY FRAGMENT HEAVY CHAIN-PROTEIN, 9D5-HEAVY CHAIN, ... | | Authors: | Aravind, P, Wang, K, Gouaux, E. | | Deposit date: | 2015-01-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Neurotransmitter and psychostimulant recognition by the dopamine transporter.
Nature, 521, 2015
|
|
1IEK
 
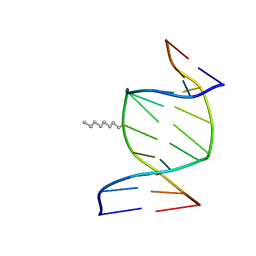 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER S) | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1J2Z
 
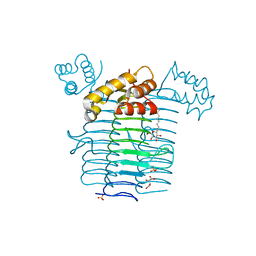 | | Crystal structure of UDP-N-acetylglucosamine acyltransferase | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, L(+)-TARTARIC ACID, SULFATE ION, ... | | Authors: | Lee, B.I, Suh, S.W. | | Deposit date: | 2003-01-15 | | Release date: | 2004-01-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of UDP-N-acetylglucosamine acyltransferase from Helicobacter pylori
Proteins, 53, 2003
|
|
1J6X
 
 | | CRYSTAL STRUCTURE OF HELICOBACTER PYLORI LUXS | | Descriptor: | AUTOINDUCER-2 PRODUCTION PROTEIN LUXS, METHIONINE, ZINC ION | | Authors: | Lewis, H.A, Furlong, E.B, Bergseid, M.G, Sanderson, W.E, Buchanan, S.G. | | Deposit date: | 2001-05-14 | | Release date: | 2001-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | A structural genomics approach to the study of quorum sensing: crystal structures of three LuxS orthologs.
Structure, 9, 2001
|
|
3UMI
 
 | | X-ray structure of the E2 domain of the human amyloid precursor protein (APP) in complex with zinc | | Descriptor: | ACETATE ION, Amyloid beta A4 protein, CADMIUM ION, ... | | Authors: | Dahms, S.O, Konnig, I, Roeser, D, Guhrs, K.H, Than, M.E. | | Deposit date: | 2011-11-13 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal Binding Dictates Conformation and Function of the Amyloid Precursor Protein (APP) E2 Domain.
J.Mol.Biol., 416, 2012
|
|
1IEY
 
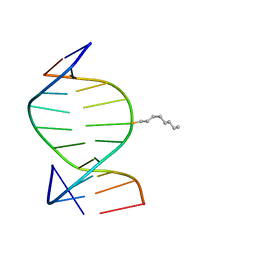 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER R) | | Descriptor: | 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', 5'-D(P*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-11 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1IYG
 
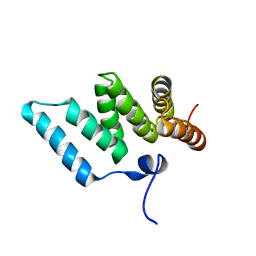 | | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA | | Descriptor: | Hypothetical protein (2010003O14) | | Authors: | Ohashi, W, Hirota, H, Yamazaki, T, Koshiba, S, Hamada, T, Yoshida, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-14 | | Release date: | 2003-02-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RSGI RUH-001, a Fis1p-like and CGI-135 homologous domain from a mouse cDNA
To be Published
|
|
1J47
 
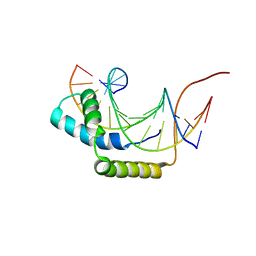 | |
