5G6Q
 
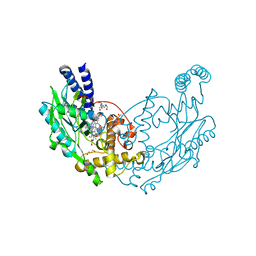 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 7-(((5-((Methylamino)methyl)pyridin-3-yl)oxy)methyl) quinolin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[5-[(methylideneamino)methyl]pyridin-3-yl]oxymethyl]quinolin-2-amine, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2016-06-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Targeting Bacterial Nitric Oxide Synthase with Aminoquinoline-Based Inhibitors.
Biochemistry, 55, 2016
|
|
5V7Y
 
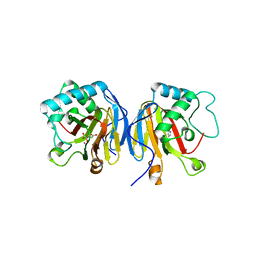 | |
5EUO
 
 | | PF6-M1-HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Yang, X. | | Deposit date: | 2015-11-18 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PF6-M1-HLA-A2 complex
To Be Published
|
|
5G3X
 
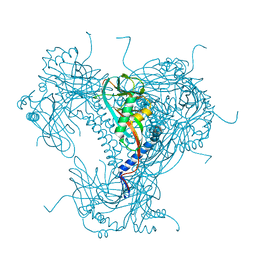 | | Structure of recombinant granulovirus polyhedrin | | Descriptor: | GRANULOVIRUS POLYHEDRIN | | Authors: | Bunker, R.D, Chiu, E, Metcalf, P. | | Deposit date: | 2016-05-02 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Atomic structure of granulin determined from native nanocrystalline granulovirus using an X-ray free-electron laser.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VB0
 
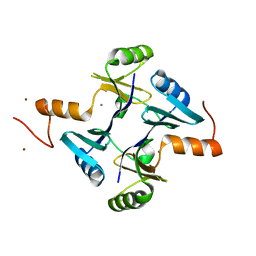 | | Crystal structure of fosfomycin resistance protein FosA3 | | Descriptor: | Fosfomycin resistance protein FosA3, MANGANESE (II) ION, NICKEL (II) ION | | Authors: | Klontz, E, Guenther, S, Silverstein, Z, Sundberg, E. | | Deposit date: | 2017-03-28 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Structure and Dynamics of FosA-Mediated Fosfomycin Resistance in Klebsiella pneumoniae and Escherichia coli.
Antimicrob. Agents Chemother., 61, 2017
|
|
8OYY
 
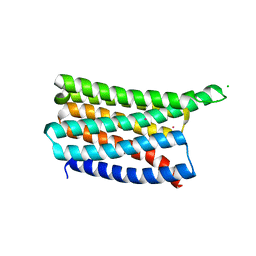 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
6WJC
 
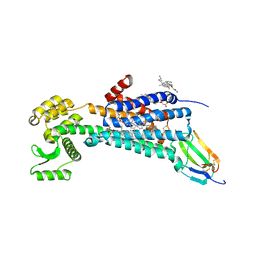 | | Muscarinic acetylcholine receptor 1 - muscarinic toxin 7 complex | | Descriptor: | (1R,5S)-8-METHYL-8-AZABICYCLO[3.2.1]OCT-3-YL (2R)-3-HYDROXY-2-PHENYLPROPANOATE, ACETAMIDE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Maeda, S, Kobilka, B.K. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure and selectivity engineering of the M1muscarinic receptor toxin complex.
Science, 369, 2020
|
|
6O0L
 
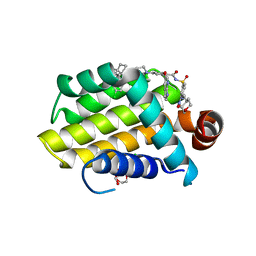 | | crystal structure of BCL-2 G101V mutation with venetoclax | | Descriptor: | 4-{4-[(4'-chloro-5,5-dimethyl[3,4,5,6-tetrahydro[1,1'-biphenyl]]-2-yl)methyl]piperazin-1-yl}-N-[(3-nitro-4-{[(oxan-4-yl )methyl]amino}phenyl)sulfonyl]-2-[(1H-pyrrolo[2,3-b]pyridin-5-yl)oxy]benzamide, Apoptosis regulator Bcl-2,Bcl-2-like protein 1,Apoptosis regulator Bcl-2, CHLORIDE ION, ... | | Authors: | Birkinshaw, R.W, Luo, C.S, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-02-16 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of BCL-2 in complex with venetoclax reveal the molecular basis of resistance mutations.
Nat Commun, 10, 2019
|
|
5VBM
 
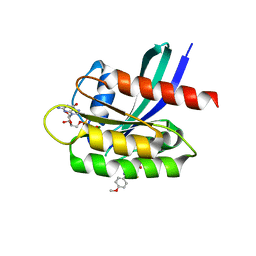 | | Crystal Structure of Small Molecule Disulfide 2C07 Bound to K-Ras Cys Light M72C GDP | | Descriptor: | 1-(4-methoxyphenyl)-N-(3-sulfanylpropyl)-5-(trifluoromethyl)-1H-pyrazole-4-carboxamide, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gentile, D.R, Jenkins, M.L, Moss, S.M, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-29 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ras Binder Induces a Modified Switch-II Pocket in GTP and GDP States.
Cell Chem Biol, 24, 2017
|
|
6ZLC
 
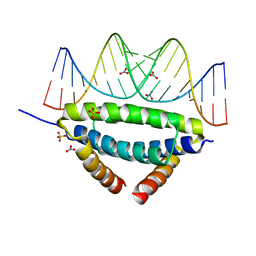 | | Non-specific dsRNA recognition by wildtype H7N1 RNA-binding domain | | Descriptor: | NITRATE ION, Non-structural protein 1, RNA (5'-R(*AP*GP*AP*CP*AP*GP*CP*AP*UP*UP*AP*UP*GP*CP*UP*GP*UP*CP*U)-3'), ... | | Authors: | Coste, F, Wacquiez, A, Marc, D, Castaing, B. | | Deposit date: | 2020-06-30 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Sequence Determinants Governing the Interactions of RNAs with Influenza A Virus Non-Structural Protein NS1.
Viruses, 12, 2020
|
|
5EV2
 
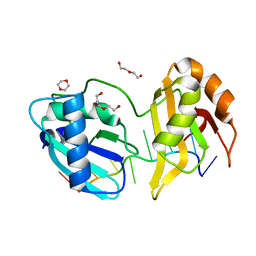 | | Structure II of Intact U2AF65 Recognizing the 3' Splice Site Signal | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, DNA (5'-R(P*UP*U)-D(P*UP*U)-R(P*U)-D(P*UP*(BRU)P*U)-3'), ... | | Authors: | Agrawal, A.A, Jenkins, J.L, Kielkopf, C.L. | | Deposit date: | 2015-11-19 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal.
Nat Commun, 7, 2016
|
|
6ZLJ
 
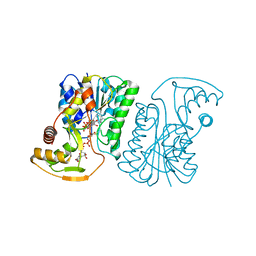 | | Crystal Structure of UDP-Glucuronic acid 4-epimerase Y149F mutant from Bacillus cereus in complex with UDP-4-DEOXY-4-FLUORO-Glucuronic acid and NAD | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{R})-6-[[[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidany l)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-3-fluoranyl-4,5-bis(oxidanyl)oxane-2-carboxylic acid, Epimerase domain-containing protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Iacovino, L.G, Mattevi, A. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic snapshots of UDP-glucuronic acid 4-epimerase ligand binding, rotation, and reduction.
J.Biol.Chem., 295, 2020
|
|
5EV3
 
 | |
5VCB
 
 | |
5GGF
 
 | |
5UJ6
 
 | |
5EUU
 
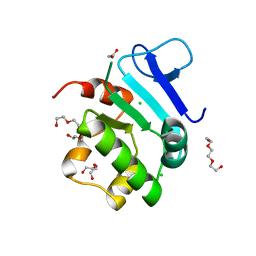 | | Rat prestin STAS domain in complex with chloride | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Prestin,Prestin, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
8ONC
 
 | |
6Z4W
 
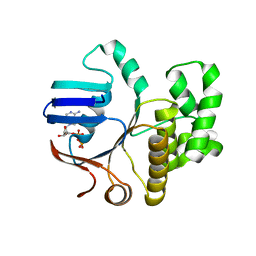 | | FtsE structure from Streptococcus pneumoniae in complex with ADP (space group P 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division ATP-binding protein FtsE | | Authors: | Alcorlo, M, Straume, D, Hermoso, J.A, Havarstein, L.S. | | Deposit date: | 2020-05-26 | | Release date: | 2020-09-02 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural Characterization of the Essential Cell Division Protein FtsE and Its Interaction with FtsX in Streptococcus pneumoniae.
Mbio, 11, 2020
|
|
8OXY
 
 | |
5EUS
 
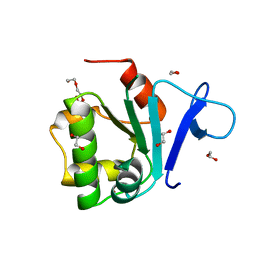 | | Rat prestin STAS domain in complex with bromide | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, Prestin,Rat prestin STAS domain, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.833 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
5EUW
 
 | | Rat prestin STAS domain in complex with nitrate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NITRATE ION, ... | | Authors: | Lolli, G, Pasqualetto, E, Costanzi, E, Bonetto, G, Battistutta, R. | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The STAS domain of mammalian SLC26A5 prestin harbours an anion-binding site.
Biochem.J., 473, 2016
|
|
6WCK
 
 | | KRAS G-quadruplex G16T mutant with Bromo Uracil replacing T8 and T16. | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*GP*GP*(BRU)P*GP*TP*GP*GP*GP*AP*AP*(BRU)P*AP*GP*GP*GP*AP*A)-3'), POTASSIUM ION | | Authors: | Schmidberger, J.W, Ou, A, Smith, N.M, Iyer, K.S, Bond, C.S. | | Deposit date: | 2020-03-30 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | High resolution crystal structure of a KRAS promoter G-quadruplex reveals a dimer with extensive poly-A pi-stacking interactions for small-molecule recognition.
Nucleic Acids Res., 48, 2020
|
|
5EV4
 
 | |
8OXV
 
 | |
