8BUH
 
 | | Structure of DDB1 bound to WX3-engaged CDK12-cyclin K | | Descriptor: | 6-[[[2-[[(2~{R})-1-oxidanylbutan-2-yl]amino]-9-propan-2-yl-purin-6-yl]amino]methyl]-3-pyridin-2-yl-1~{H}-pyridin-2-one, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.79 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8B4T
 
 | |
8BUC
 
 | | Structure of DDB1 bound to dCeMM3-engaged CDK12-cyclin K | | Descriptor: | 2-(1~{H}-benzimidazol-2-ylsulfanyl)-~{N}-(5-chloranylpyridin-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.85 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8B4W
 
 | | X-ray structure of furin (PCSK3) in complex with 1H-isoindol-3-amine | | Descriptor: | 1H-isoindol-3-amine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8BUT
 
 | | Structure of DDB1 bound to DS61-engaged CDK12-cyclin K | | Descriptor: | 2-[[6-[[4-(2-hydroxyethyloxy)phenyl]methylamino]-9-propan-2-yl-purin-2-yl]amino]butan-1-ol, Cyclin-K, Cyclin-dependent kinase 12, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8B50
 
 | |
8B4V
 
 | | X-ray structure of furin (PCSK3) in complex with benzamidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, CALCIUM ION, ... | | Authors: | Dahms, S.O, Brandstetter, H. | | Deposit date: | 2022-09-21 | | Release date: | 2023-10-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fragment-Based Design, Synthesis, and Characterization of Aminoisoindole-Derived Furin Inhibitors.
Chemmedchem, 19, 2024
|
|
8B5F
 
 | |
8BUD
 
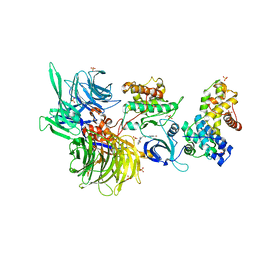 | | Structure of DDB1 bound to Z7-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUE
 
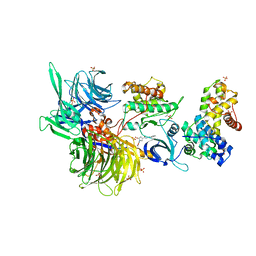 | | Structure of DDB1 bound to Z11-engaged CDK12-cyclin K | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Kozicka, Z, Kempf, G, Petzold, G, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-03 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8BUG
 
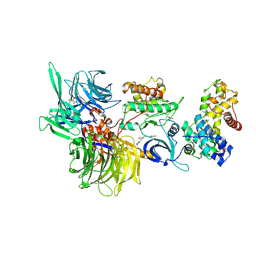 | | Structure of DDB1 bound to HQ461-engaged CDK12-cyclin K | | Descriptor: | 2-[2-[(6-methylpyridin-2-yl)amino]-1,3-thiazol-4-yl]-~{N}-(5-methyl-1,3-thiazol-2-yl)ethanamide, CITRIC ACID, Cyclin-K, ... | | Authors: | Kozicka, Z, Kempf, G, Focht, V, Thoma, N.H. | | Deposit date: | 2022-11-30 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Design principles for cyclin K molecular glue degraders.
Nat.Chem.Biol., 20, 2024
|
|
8B9N
 
 | |
8B66
 
 | |
8BE4
 
 | |
8BAP
 
 | | Eugenol Oxidase (EUGO) from Rhodococcus jostii RHA1, eightfold mutant active on propanol syringol | | Descriptor: | 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2,6-dimethoxyphenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-10-11 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | One-Pot Biocatalytic Synthesis of rac -Syringaresinol from a Lignin-Derived Phenol.
Acs Catalysis, 13, 2023
|
|
8BCM
 
 | |
8BA7
 
 | |
8SS3
 
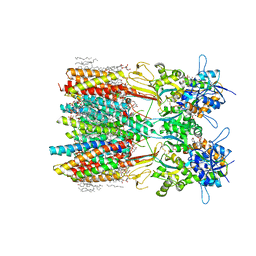 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.21 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SSA
 
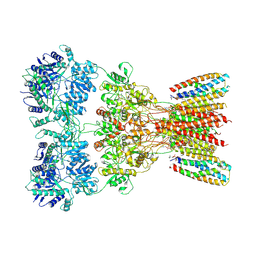 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to glutamate and channel blocker spermidine (desensitized state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, GLUTAMIC ACID, Glutamate receptor 2, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS7
 
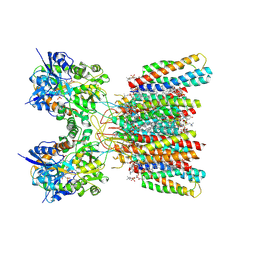 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8SS6
 
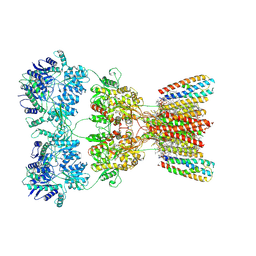 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK, channel blocker spermidine and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GET
 
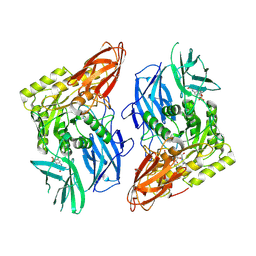 | | R. hominis 2 beta-glucuronidase bound to norquetiapine-glucuronide | | Descriptor: | 11-(4-beta-D-glucopyranuronosylpiperazin-1-yl)dibenzo[b,f][1,4]thiazepine, FLAVIN MONONUCLEOTIDE, GLYCEROL, ... | | Authors: | Simpson, J.B, Redinbo, M.R. | | Deposit date: | 2023-03-07 | | Release date: | 2024-03-20 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Gut microbial beta-glucuronidases influence endobiotic homeostasis and are modulated by diverse therapeutics.
Cell Host Microbe, 32, 2024
|
|
8SS2
 
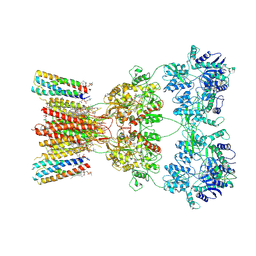 | | Structure of AMPA receptor GluA2 complex with auxiliary subunits TARP gamma-5 and cornichon-2 bound to competitive antagonist ZK and channel blocker spermidine (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, Digitonin, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-01 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8GKF
 
 | |
8SS9
 
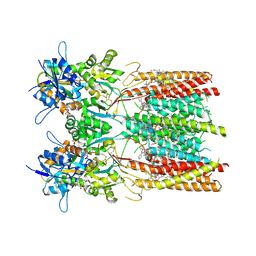 | | Structure of LBD-TMD of AMPA receptor GluA2 in complex with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK and antiepileptic drug perampanel (closed state) | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-(6'-oxo-1'-phenyl[1',6'-dihydro[2,3'-bipyridine]]-5'-yl)benzonitrile, CHOLESTEROL, ... | | Authors: | Gangwar, S.P, Yen, L.Y, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2023-05-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Modulation of GluA2-gamma 5 synaptic complex desensitization, polyamine block and antiepileptic perampanel inhibition by auxiliary subunit cornichon-2.
Nat.Struct.Mol.Biol., 30, 2023
|
|
