5B4O
 
 | | Crystal structure of Macrophage Migration Inhibitory Factor in complex with BTZO-14 | | Descriptor: | 1,2-ETHANEDIOL, 2-pyridin-3-yl-1,3-benzothiazin-4-one, Macrophage migration inhibitory factor, ... | | Authors: | Oki, H, Igaki, S, Moriya, Y, Hayano, Y, Habuka, N. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | BTZO-1, a cardioprotective agent, reveals that macrophage migration inhibitory factor regulates ARE-mediated gene expression
Chem. Biol., 17, 2010
|
|
5BRY
 
 | | HIV-1 wild Type protease with GRL-011-11A (a methylamine bis-Tetrahydrofuran P2-Ligand, sulfonamide isostere derivate) | | Descriptor: | (3R,3aS,4R,6aR)-4-(methylamino)hexahydrofuro[2,3-b]furan-3-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with Amino-bis-tetrahydrofuran Derivatives as P2-Ligands to Enhance Backbone-Binding Interactions: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Studies.
J.Med.Chem., 58, 2015
|
|
3JSM
 
 | | K65R mutant HIV-1 reverse transcriptase cross-linked to DS-DNA and complexed with tenofovir-diphosphate as the incoming nucleotide substrate | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(DDG))-3'), DNA (5'-D(*A*TP*GP*GP*TP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), HIV-1 REVERSE TRANSCRIPTASE P51 SUBUNIT, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2009-09-10 | | Release date: | 2009-09-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the role of the K65r mutation in HIV-1 reverse transcriptase polymerization, excision antagonism, and tenofovir resistance.
J.Biol.Chem., 284, 2009
|
|
4HO1
 
 | |
7JMD
 
 | | Sheep Connexin-46 at 2.5 angstroms resolution, Lipid Class 1 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-3 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.A, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
2XEY
 
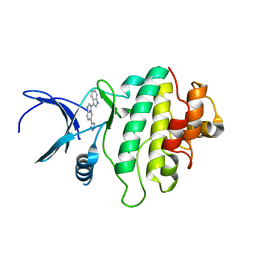 | | Crystal structure of checkpoint kinase 1 (Chk1) in complex with inhibitors | | Descriptor: | 3-(1H-INDOL-3-YL)-6-(1H-PYRAZOL-4-YL)IMIDAZO[1,2-A]PYRAZINE, SERINE/THREONINE-PROTEIN KINASE CHK1 | | Authors: | Matthews, T.P, McHardy, T, Klair, S, Boxall, K, Fisher, M, Cherry, M, Allen, C.E, Addison, G.J, Ellard, J, Aherne, G.W, Westwood, I.M, van Montfort, R, Garrett, M.D, Reader, J.C, Collins, I. | | Deposit date: | 2010-05-19 | | Release date: | 2010-06-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and Evaluation of 3,6-Di(Hetero)Aryl Imidazo[1,2-A]Pyrazines as Inhibitors of Checkpoint and Other Kinases.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4DBN
 
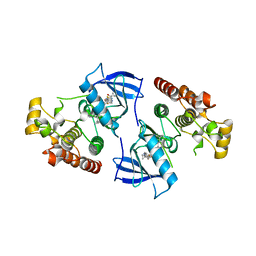 | | Crystal Structure of the Kinase domain of Human B-raf with a [1,3]thiazolo[5,4-b]pyridine derivative | | Descriptor: | 2-chloro-3-(1-cyanocyclopropyl)-N-[5-({2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}oxy)-2-fluorophenyl]benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2012-01-16 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors. 1. Exploration of [5,6]-fused bicyclic scaffolds.
J.Med.Chem., 55, 2012
|
|
4DJP
 
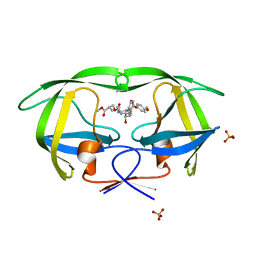 | | Crystal Structure of wild-type HIV-1 Protease in Complex with MKP73 | | Descriptor: | PHOSPHATE ION, Pol polyprotein, methyl (2S)-3-({[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl][(2S)-2-methylbutyl]amino}-1-phenylbutan-2-yl]carbamoyl}oxy)-2-methylpropanoate | | Authors: | Schiffer, C.A, Nalam, M.N.L. | | Deposit date: | 2012-02-02 | | Release date: | 2012-08-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance.
J.Med.Chem., 55, 2012
|
|
3CV0
 
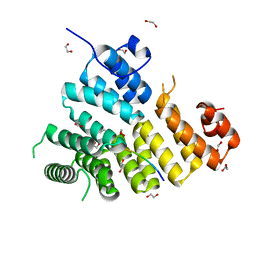 | | Structure of Peroxisomal Targeting Signal 1 (PTS1) binding domain of Trypanosoma brucei Peroxin 5 (TbPEX5)complexed to T. brucei Phosphoglucoisomerase (PGI) PTS1 peptide | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome targeting signal 1 receptor PEX5, T. brucei PGI PTS1 peptide Ac-FNELSHL | | Authors: | Sampathkumar, P, Roach, C, Michels, P.A.M, Hol, W.G.J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-06-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into the recognition of peroxisomal targeting signal 1 by Trypanosoma brucei peroxin 5.
J.Mol.Biol., 381, 2008
|
|
7VIG
 
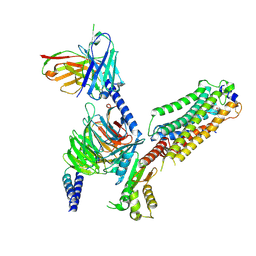 | | Cryo-EM structure of Gi coupled Sphingosine 1-phosphate receptor bound with CBP-307 | | Descriptor: | 1-[[2-fluoranyl-4-[5-[4-(2-methylpropyl)phenyl]-1,2,4-oxadiazol-3-yl]phenyl]methyl]azetidine-3-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yu, L.Y, Gan, B, Xiao, Q.J, Ren, R.B. | | Deposit date: | 2021-09-26 | | Release date: | 2022-09-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural insights into sphingosine-1-phosphate receptor activation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3HH3
 
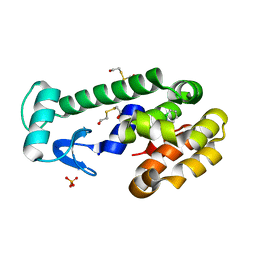 | | New azaborine compounds bind to the T4 lysozyme L99A cavity - 1,2-dihydro-1,2-azaborine | | Descriptor: | 1,2-dihydro-1,2-azaborinine, 2-HYDROXYETHYL DISULFIDE, Lysozyme, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2009-05-14 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Boron mimetics: 1,2-dihydro-1,2-azaborines bind inside a nonpolar cavity of T4 lysozyme.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4XZ2
 
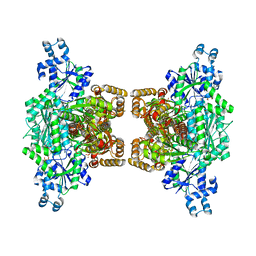 | | Human platelet phosphofructokinase in an R-state in complex with ADP and F6P, crystal form I | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kloos, M, Strater, N. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Crystal structure of human platelet phosphofructokinase-1 locked in an activated conformation.
Biochem.J., 469, 2015
|
|
3HLJ
 
 | | Crystal structure of human carbonic anhydrase isozyme II with 3-methylthiobenzimidazo[1,2-c][1,2,3]thiadiazol-7-sulfonamide | | Descriptor: | 3-methylthiobenzimidazo[1,2-c][1,2,3]thiadiazol-7-sulfonamide, Carbonic anhydrase 2, SODIUM ION, ... | | Authors: | Grazulis, S, Manakova, E, Golovenko, D. | | Deposit date: | 2009-05-27 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Inhibition and binding studies of carbonic anhydrase isozymes I, II and IX with benzimidazo[1,2-c][1,2,3]thiadiazole-7-sulphonamides
J Enzyme Inhib Med Chem, 25, 2010
|
|
4O0Y
 
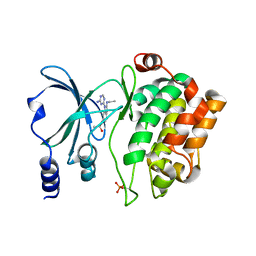 | | Back pocket flexibility provides group-II PAK selectivity for type 1 kinase inhibitors | | Descriptor: | 4-[1-(4-amino-1,3,5-triazin-2-yl)-2-(ethylamino)-1H-benzimidazol-6-yl]-2-methylbut-3-yn-2-ol, Serine/threonine-protein kinase PAK 4 | | Authors: | Rouge, L, Tam, C, Wang, W. | | Deposit date: | 2013-12-14 | | Release date: | 2014-02-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Back Pocket Flexibility Provides Group II p21-Activated Kinase (PAK) Selectivity for Type I 1/2 Kinase Inhibitors.
J.Med.Chem., 57, 2014
|
|
3VBQ
 
 | |
3I6O
 
 | | Crystal structure of wild type HIV-1 protease with macrocyclic inhibitor GRL-0216A | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(1S,2R)-1-benzyl-2-hydroxy-3-[(7E)-13-methoxy-1,1-dioxido-3,4,5,6,9,10-hexahydro-2H-11,1,2-benzoxathiazacyclotridecin-2-yl]propyl}carbamate, GLYCEROL, IODIDE ION, ... | | Authors: | Chumanevich, A.A, Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2009-07-07 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Design, Synthesis, Protein-Ligand X-ray Structure, and Biological Evaluation of a Series of Novel Macrocyclic Human Immunodeficiency Virus-1 Protease Inhibitors to Combat Drug Resistance.
J.Med.Chem., 52, 2009
|
|
4ZAC
 
 | | Structure of S. cerevisiae Fdc1 with the prenylated-flavin cofactor in the iminium form. | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | White, M.D, Leys, D. | | Deposit date: | 2015-04-13 | | Release date: | 2015-06-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | New cofactor supports alpha , beta-unsaturated acid decarboxylation via 1,3-dipolar cycloaddition.
Nature, 522, 2015
|
|
4JEC
 
 | | Joint neutron and X-ray structure of per-deuterated HIV-1 protease in complex with clinical inhibitor amprenavir | | Descriptor: | CHLORIDE ION, HIV-1 protease, {3-[(4-AMINO-BENZENESULFONYL)-ISOBUTYL-AMINO]-1-BENZYL-2-HYDROXY-PROPYL}-CARBAMIC ACID TETRAHYDRO-FURAN-3-YL ESTER | | Authors: | Kovalevsky, A.Y, Weber, I.T, Langan, P. | | Deposit date: | 2013-02-26 | | Release date: | 2013-07-24 | | Last modified: | 2025-11-12 | | Method: | NEUTRON DIFFRACTION (2.01 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/Neutron Crystallographic Study of HIV-1 Protease with Clinical Inhibitor Amprenavir: Insights for Drug Design.
J.Med.Chem., 56, 2013
|
|
3IFC
 
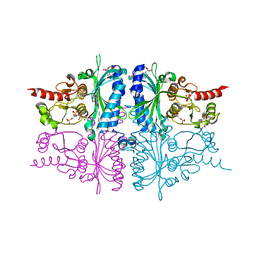 | | Human muscle fructose-1,6-bisphosphatase E69Q mutant in complex with AMP and alpha fructose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-fructofuranose, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase isozyme 2, ... | | Authors: | Kolodziejczyk, R, Zarzycki, M, Jaskolski, M, Dzugaj, A. | | Deposit date: | 2009-07-24 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure of E69Q mutant of human muscle fructose-1,6-bisphosphatase
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4NFM
 
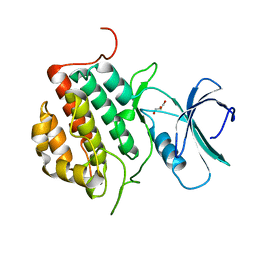 | | Human tau tubulin kinase 1 (TTBK1) | | Descriptor: | GLYCEROL, Tau-tubulin kinase 1 | | Authors: | Sheriff, S. | | Deposit date: | 2013-10-31 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The structure of human tau-tubulin kinase 1 both in the apo form and in complex with an inhibitor.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4ZIP
 
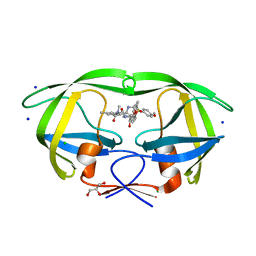 | | HIV-1 wild Type protease with GRL-0648A (a isophthalamide-derived P2-Ligand) | | Descriptor: | CHLORIDE ION, GLYCEROL, N-[(2,5-dimethyl-1,3-oxazol-4-yl)methyl]-N'-[(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]-N,5-dimethylbenzene-1,3-dicarboxamide, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-04-28 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Structure-based design, synthesis, X-ray studies, and biological evaluation of novel HIV-1 protease inhibitors containing isophthalamide-derived P2-ligands.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4ZLS
 
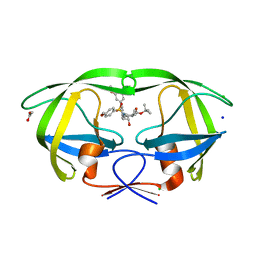 | | HIV-1 wild Type protease with GRL-096-13A (a Boc-derivative P2-Ligand, 3,-5-dimethylbiphenyl P1-Ligand) | | Descriptor: | ACETATE ION, CHLORIDE ION, Protease, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2015-05-01 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structure-Based Design of Potent HIV-1 Protease Inhibitors with Modified P1-Biphenyl Ligands: Synthesis, Biological Evaluation, and Enzyme-Inhibitor X-ray Structural Studies.
J.Med.Chem., 58, 2015
|
|
4AIE
 
 | | Structure of glucan-1,6-alpha-glucosidase from Lactobacillus acidophilus NCFM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLUCAN 1,6-ALPHA-GLUCOSIDASE, ... | | Authors: | Fredslund, F, Navarro Poulsen, J.C, Lo Leggio, L. | | Deposit date: | 2012-02-09 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymology and Structure of the Gh13_31 Glucan 1,6-Alpha-Glucosidase that Confers Isomaltooligosaccharide Utilization in the Probiotic Lactobacillus Acidophilus Ncfm.
J.Bacteriol., 194, 2012
|
|
