5NMP
 
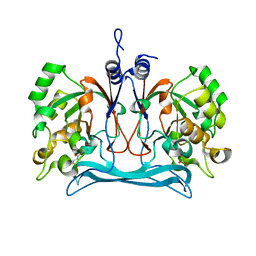 | |
5NLF
 
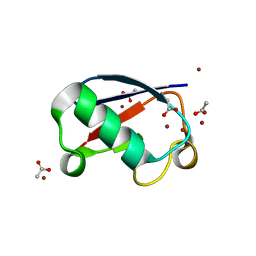 | | Crystal structure of Zn2.7-E16V human ubiquitin (hUb) mutant adduct, from a solution 100 mM zinc acetate/1.3 mM E16V hUb | | Descriptor: | ACETATE ION, Polyubiquitin-C, ZINC ION | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
5NLJ
 
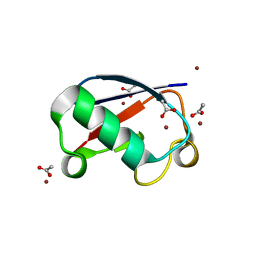 | | Crystal structure of Zn3-E16V human ubiquitin (hUb) mutant adduct, from a solution 70 mM zinc acetate/20% v/v TFE/1.3 mM E16V hUb | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fermani, S, Falini, G. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Aggregation Pathways of Native-Like Ubiquitin Promoted by Single-Point Mutation, Metal Ion Concentration, and Dielectric Constant of the Medium.
Chemistry, 24, 2018
|
|
3C13
 
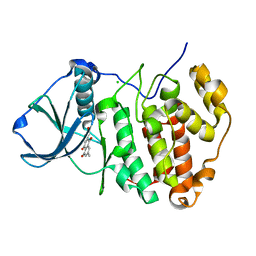 | | Low pH-value crystal structure of emodin in complex with the catalytic subunit of protein kinase CK2 | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, CHLORIDE ION, Casein kinase II subunit alpha | | Authors: | Niefind, K, Raaf, J, Issinger, O.-G. | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Catalytic Subunit of Human Protein Kinase CK2 Structurally Deviates from Its Maize Homologue in Complex with the Nucleotide Competitive Inhibitor Emodin
J.Mol.Biol., 377, 2008
|
|
1KX9
 
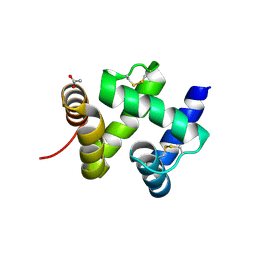 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
2F19
 
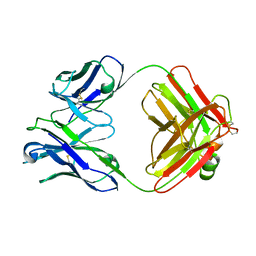 | | THREE-DIMENSIONAL STRUCTURE OF TWO CRYSTAL FORMS OF FAB R19.9, FROM A MONOCLONAL ANTI-ARSONATE ANTIBODY | | Descriptor: | IGG2B-KAPPA R19.9 FAB (HEAVY CHAIN), IGG2B-KAPPA R19.9 FAB (LIGHT CHAIN) | | Authors: | Lascombe, M.B, Alzari, P.M, Poljak, R.J, Nisonoff, A. | | Deposit date: | 1992-05-27 | | Release date: | 1992-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Three-dimensional structure of two crystal forms of FabR19.9 from a monoclonal anti-arsonate antibody.
Proc.Natl.Acad.Sci.USA, 89, 1992
|
|
6I8H
 
 | |
6ILI
 
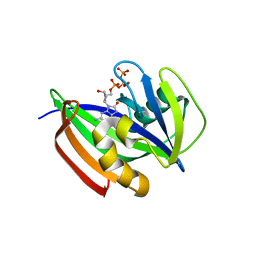 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
5NFR
 
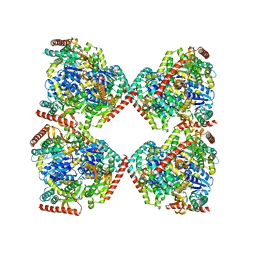 | | Crystal structure of malate dehydrogenase from Plasmodium falciparum (PfMDH) | | Descriptor: | CITRIC ACID, Malate dehydrogenase | | Authors: | Lunev, S, Romero, A.R, Batista, F.A, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-03-15 | | Release date: | 2018-01-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oligomeric interfaces as a tool in drug discovery: Specific interference with activity of malate dehydrogenase of Plasmodium falciparum in vitro.
PLoS ONE, 13, 2018
|
|
1I0I
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-GLUTAMINE MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-29 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
1I0L
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-ASPARAGINE MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-29 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
6HR2
 
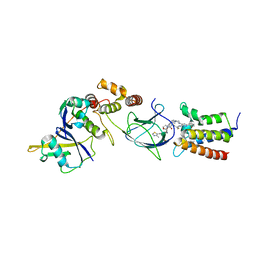 | | Crystal structure of PROTAC 2 in complex with the bromodomain of human SMARCA4 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[4-[[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]methyl]phenyl]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
1I13
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-ALANINE MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, FORMIC ACID, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-30 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
1I14
 
 | | ANALYSIS OF AN INVARIANT ASPARTIC ACID IN HPRTS-GLUTAMIC ACID MUTANT | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 7-HYDROXY-PYRAZOLO[4,3-D]PYRIMIDINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Canyuk, B, Focia, P.J, Eakin, A.E. | | Deposit date: | 2001-01-30 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role for an invariant aspartic acid in hypoxanthine phosphoribosyltransferases is examined using saturation mutagenesis, functional analysis, and X-ray crystallography.
Biochemistry, 40, 2001
|
|
6HX5
 
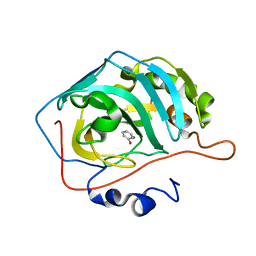 | |
1KP7
 
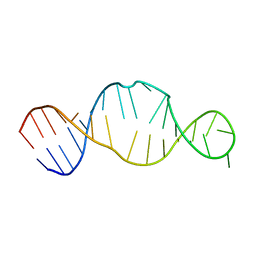 | | Conserved RNA Structure within the HCV IRES eIF3 Binding Site | | Descriptor: | Hepatitis C Virus Internal Ribosome Entry Site Fragment | | Authors: | Gallego, J, Klinck, R, Collier, A.J, Cole, P.T, Harris, S.J, Harrison, G.P, Aboul-ela, F, Walker, S, Varani, G. | | Deposit date: | 2001-12-29 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A conserved RNA structure within the HCV IRES eIF3-binding site.
Nat.Struct.Biol., 9, 2002
|
|
1KHI
 
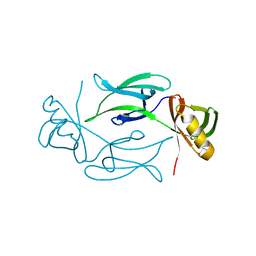 | | CRYSTAL STRUCTURE OF HEX1 | | Descriptor: | Hex1 | | Authors: | Yuan, P, Swaminathan, K. | | Deposit date: | 2001-11-30 | | Release date: | 2002-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa
NAT.STRUCT.BIOL., 10, 2003
|
|
8EVD
 
 | |
5M91
 
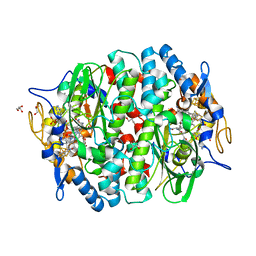 | | PCE reductive dehalogenase from S. multivorans in complex with 2,6-dibromophenol | | Descriptor: | 2,6-bis(bromanyl)phenol, BENZAMIDINE, GLYCEROL, ... | | Authors: | Kunze, C, Bommer, M, Hagen, W.R, Uksa, M, Dobbek, H, Schubert, T, Diekert, G. | | Deposit date: | 2016-10-31 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Cobamide-mediated enzymatic reductive dehalogenation via long-range electron transfer.
Nat Commun, 8, 2017
|
|
5M66
 
 | |
6SNP
 
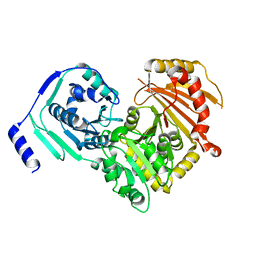 | | Crystal structures of human PGM1 isoform 2 | | Descriptor: | MAGNESIUM ION, Phosphoglucomutase-1 | | Authors: | Backe, P.H, Laerdahl, J.K, Kittelsen, L.S, Dalhus, B, Morkrid, L, Bjoras, M. | | Deposit date: | 2019-08-27 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for substrate and product recognition in human phosphoglucomutase-1 (PGM1) isoform 2, a member of the alpha-D-phosphohexomutase superfamily.
Sci Rep, 10, 2020
|
|
6HFI
 
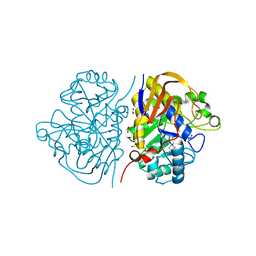 | |
6HQA
 
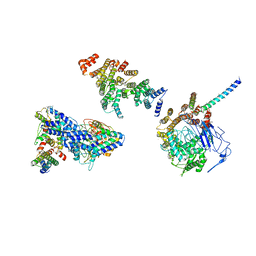 | | Molecular structure of promoter-bound yeast TFIID | | Descriptor: | Histone-fold, Subunit (17 kDa) of TFIID and SAGA complexes, involved in RNA polymerase II transcription initiation, ... | | Authors: | Kolesnikova, O, Ben-Shem, A, Luo, J, Ranish, J, Schultz, P, Papai, G. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Molecular structure of promoter-bound yeast TFIID.
Nat Commun, 9, 2018
|
|
5LIP
 
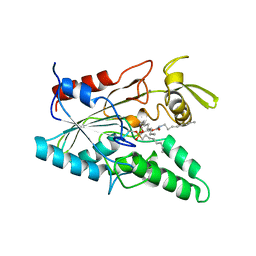 | | PSEUDOMONAS LIPASE COMPLEXED WITH RC-(RP, SP)-1,2-DIOCTYLCARBAMOYLGLYCERO-3-O-OCTYLPHOSPHONATE | | Descriptor: | CALCIUM ION, OCTYL-PHOSPHINIC ACID 1,2-BIS-OCTYLCARBAMOYLOXY-ETHYL ESTER, TRIACYL-GLYCEROL HYDROLASE | | Authors: | Lang, D.A, Dijkstra, B.W. | | Deposit date: | 1997-09-02 | | Release date: | 1998-08-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of the chiral selectivity of Pseudomonas cepacia lipase
Eur.J.Biochem., 254, 1998
|
|
6FQF
 
 | | THE X-RAY STRUCTURE OF FERRIC-BETA4 HUMAN HEMOGLOBIN | | Descriptor: | Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Merlino, A, Balasco, N, Balsamo, A, Vergara, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ferric homotetrameric beta4human hemoglobin.
Biophys. Chem., 240, 2018
|
|
