8PKO
 
 | | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer). | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Roversi, P, Hitchman, C.J, Lia, A, Bayo, Y. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | The ERAD misfolded glycoprotein checkpoint complex from Chaetomium thermophilum (EDEM:PDI heterodimer).
To Be Published
|
|
8PMG
 
 | | Structure of Nal1 indica cultivar IR64, construct 36-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMI
 
 | | Structure of Nal1 indica cultivar IR64, construct 36-458 in presence of peptide from FZP protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, AMMONIUM ION, MAGNESIUM ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-28 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PML
 
 | | Structure of Nal1 protein , SPIKE allele from japonica rice, construct 46-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PMM
 
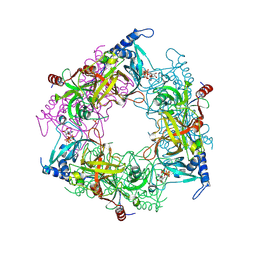 | | Structure of Nal1 protein, allele SPIKE from japonica rice, construct 31-458 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PN1
 
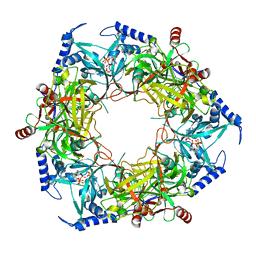 | | CryoEM structure of Nal1 protein, allele SPIKE, from Oryza sativa japonica group | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PN2
 
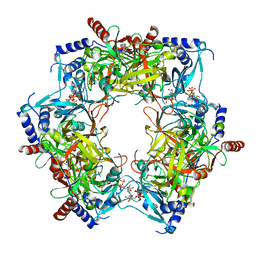 | | CryoEM structure of Nal1 protein, allele IR64, from Oryza sativa indica cultivar | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein NARROW LEAF 1 | | Authors: | Huang, L.Y, Rety, S, Xi, X.G. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-17 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | The catalytic triad of rice NARROW LEAF1 involves H234.
Nat.Plants, 10, 2024
|
|
8PPK
 
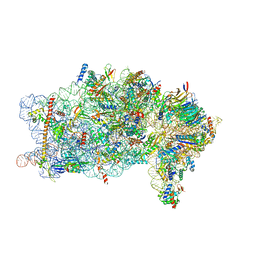 | | Bat-Hp-CoV Nsp1 and eIF1 bound to the human 40S small ribosomal subunit | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8PPL
 
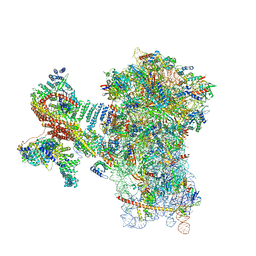 | | MERS-CoV Nsp1 bound to the human 43S pre-initiation complex | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Ban, I, Lapointe, C.P, Leibundgut, M, Baeumlin, E, Kummerant, E, Scaiola, A, Schoenhut, T, Ziegelmueller, J, Puglisi, J.D, Muehlemann, O, Ban, N. | | Deposit date: | 2023-07-07 | | Release date: | 2023-10-18 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Universal features of Nsp1-mediated translational shutdown by coronaviruses.
Mol.Cell, 83, 2023
|
|
8Q5I
 
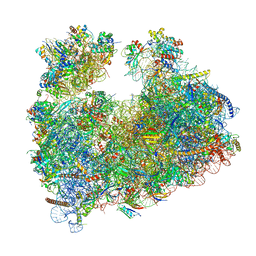 | | Structure of Candida albicans 80S ribosome in complex with cephaeline | | Descriptor: | 18S ribosomal RNA, 25S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kolosova, O, Zgadzay, Y, Stetsenko, A, Atamas, A, Guskov, A, Yusupov, M. | | Deposit date: | 2023-08-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Structural characterization of cephaeline binding to the eukaryotic ribosome using Cryo-Electron Microscopy
Biopolym Cell, 2023
|
|
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 2024
|
|
8R6F
 
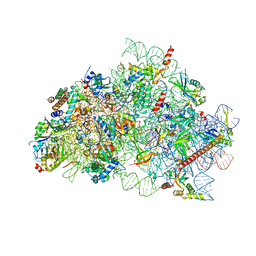 | | CryoEM structure of wheat 40S ribosomal subunit, body domain | | Descriptor: | 30S ribosomal protein S11, chloroplastic, 30S ribosomal protein S4, ... | | Authors: | Kravchenko, O.V, Baymukhametov, T.N, Afonina, Z.A, Vasilenko, K.S. | | Deposit date: | 2023-11-22 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | High-Resolution Structure and Internal Mobility of a Plant 40S Ribosomal Subunit.
Int J Mol Sci, 24, 2023
|
|
8RXH
 
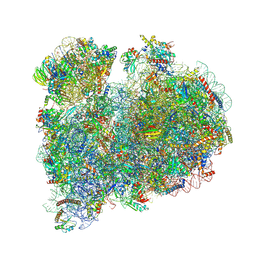 | | CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN | | Descriptor: | (2S)-2-[2-[4-[[(2R,3S,4S)-3-acetyloxy-4-oxidanyl-pyrrolidin-2-yl]methyl]phenoxy]ethanoylamino]-6-azanyl-hexanoic acid, 40S ribosomal protein S12, 40S ribosomal protein S14, ... | | Authors: | Rajan, K.S, Yonath, A. | | Deposit date: | 2024-02-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification.
Cell Rep, 43, 2024
|
|
8RXX
 
 | |
8S9Y
 
 | | Taipan Natriuretic Peptide C -TNPc | | Descriptor: | Peptide TNP-c | | Authors: | Torres, A.M, Alewood, P.F. | | Deposit date: | 2023-03-30 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Taipan Natriuretic Peptides Are Potent and Selective Agonists for the Natriuretic Peptide Receptor A.
Molecules, 28, 2023
|
|
8SCB
 
 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure and consequences of eRF1 glued to the ribosomal decoding center
To Be Published
|
|
8SFS
 
 | | High Affinity nanobodies against GFP | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonnano, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFV
 
 | | High affinity nanobodies to GFP | | Descriptor: | GLYCEROL, Green fluorescent protein, LaG19, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | High affinity nanobodies to GFP
To Be Published
|
|
8SFX
 
 | | High Affinity nanobodies against GFP | | Descriptor: | D-MALATE, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SFZ
 
 | | High Affinity nanobodies against GFP | | Descriptor: | Green fluorescent protein, LaG35, POTASSIUM ION, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SG3
 
 | |
8SLC
 
 | | High Affinity nanobodies against GFP | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Ketaren, N.E, Rout, M.P, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2023-04-21 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | High Affinity nanobodies against GFP
To Be Published
|
|
8SMU
 
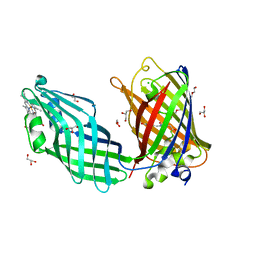 | | Integral fusion of the HtaA CR2 domain from Corynebacterium diphtheriae within EGFP | | Descriptor: | CHLORIDE ION, GLYCEROL, HtaACR2 integral fusion within enhanced green fluorescent protein, ... | | Authors: | Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2023-04-26 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Development and atomic structure of a new fluorescence-based sensor to probe heme transfer in bacterial pathogens.
J.Inorg.Biochem., 249, 2023
|
|
8SOQ
 
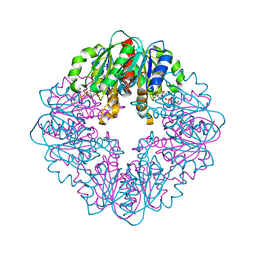 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
8STC
 
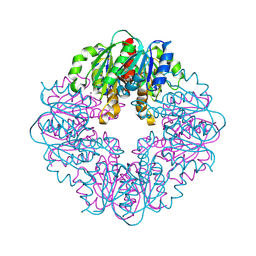 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zinc and soaked with bicarbonate. | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zinc and soaked with bicarbonate.
To be published
|
|
