8U4R
 
 | |
5M35
 
 | |
1M8X
 
 | | CRYSTAL STRUCTURE OF THE PUMILIO-HOMOLOGY DOMAIN FROM HUMAN PUMILIO1 IN COMPLEX WITH NRE1-14 RNA | | Descriptor: | 5'-R(P*UP*GP*UP*AP*UP*AP*U)-3', 5'-R(P*UP*UP*GP*UP*AP*UP*AP*U)-3', Pumilio 1 | | Authors: | Wang, X, McLachlan, J, Zamore, P.D, Hall, T.M.T. | | Deposit date: | 2002-07-26 | | Release date: | 2002-09-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MODULAR RECOGNITION OF RNA BY A HUMAN PUMILIO-HOMOLOGY DOMAIN
CELL(CAMBRIDGE,MASS.), 110, 2002
|
|
1M6O
 
 | | Crystal Structure of HLA B*4402 in complex with HLA DPA*0201 peptide | | Descriptor: | Beta-2-microglobulin, HLA DPA*0201 peptide, HLA class I histocompatibility antigen, ... | | Authors: | Macdonald, W.A, Purcell, A.W, Williams, D.S, Mifsud, N.A, Ely, L.K, Gorman, J.J, Clements, C.S, Kjer-Nielsen, L, Koelle, D.M, Brooks, A.G, Lovrecz, G.O, Lu, L, Rossjohn, J, McCluskey, J. | | Deposit date: | 2002-07-17 | | Release date: | 2003-09-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition.
J.Exp.Med., 198, 2003
|
|
1M9P
 
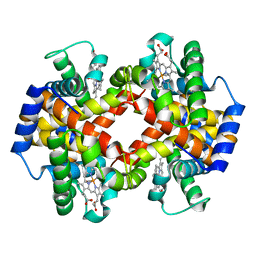 | | Crystalline Human Carbonmonoxy Hemoglobin C Exhibits The R2 Quaternary State at Neutral pH In The Presence of Polyethylene Glycol: The 2.1 Angstrom Resolution Crystal Structure | | Descriptor: | CARBON MONOXIDE, Hemoglobin alpha chain, Hemoglobin beta chain, ... | | Authors: | Patskovska, L.N, Patskovsky, Y.V, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2002-07-29 | | Release date: | 2003-08-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | COHbC and COHbS crystallize in the R2 quaternary state at neutral pH in the presence of PEG 4000.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1M6P
 
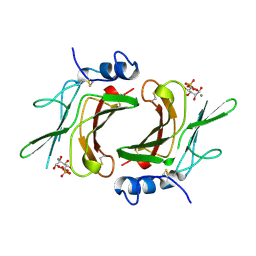 | | EXTRACYTOPLASMIC DOMAIN OF BOVINE CATION-DEPENDENT MANNOSE 6-PHOSPHATE RECEPTOR | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, CATION-DEPENDENT MANNOSE-6-PHOSPHATE RECEPTOR, MANGANESE (II) ION | | Authors: | Roberts, D.L, Weix, D.J, Dahms, N.M, Kim, J.J.-P. | | Deposit date: | 1998-04-19 | | Release date: | 1999-04-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of lysosomal enzyme recognition: three-dimensional structure of the cation-dependent mannose 6-phosphate receptor.
Cell(Cambridge,Mass.), 93, 1998
|
|
8TG5
 
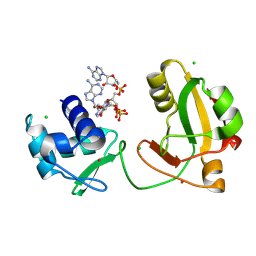 | | tRNA 2'-phosphotransferase (Tpt1) from Pyrococcus horikoshii in complex with branched 2'-PO4 RNA | | Descriptor: | (2S,3R,4R,5S)-2-(6-amino-9H-purin-9-yl)-4-{[(S)-{[(2R,3S,4R,5S)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methoxy}(hydroxy)phosphoryl]oxy}-5-({[(S)-{[(2R,3R,4S,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-2-(hydroxymethyl)oxolan-3-yl]oxy}(hydroxy)phosphoryl]oxy}methyl)oxolan-3-yl dihydrogen phosphate (non-preferred name), CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Jacewicz, A, Dantuluri, S, Shuman, S. | | Deposit date: | 2023-07-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for Tpt1-catalyzed 2'-PO 4 transfer from RNA and NADP(H) to NAD.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1W4U
 
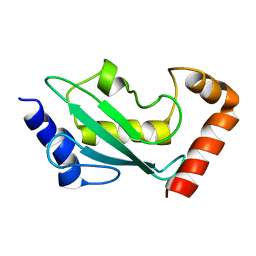 | | NMR solution structure of the ubiquitin conjugating enzyme UbcH5B | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2-17 KDA 2 | | Authors: | Houben, K, Dominguez, C, Van Schaik, F.M.A, Timmers, H.T.M, Bonvin, A.M.J.J, Boelens, R. | | Deposit date: | 2004-07-29 | | Release date: | 2004-11-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Ubiquitin-Conjugating Enzyme Ubch5B
J.Mol.Biol., 344, 2004
|
|
1W4Y
 
 | | Ferrous horseradish peroxidase C1A in complex with carbon monoxide | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, HORSERADISH PEROXIDASE C1A, ... | | Authors: | Carlsson, G.H, Nicholls, P, Svistunenko, D, Berglund, G.I, Hajdu, J. | | Deposit date: | 2004-08-03 | | Release date: | 2005-01-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Complexes of Horseradish Peroxidase with Formate, Acetate, and Carbon Monoxide
Biochemistry, 44, 2005
|
|
2FZH
 
 | | New Insights into Dihydrofolate Reductase Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
8TPX
 
 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 3 in complex with antibody fragment 1B2: trans-oriented 1B2 and ACP | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 2024
|
|
8TPW
 
 | | Crosslinked 6-deoxyerythronolide B synthase (DEBS) Module 3 in complex with antibody fragment 1B2: cis-oriented 1B2 and ACP | | Descriptor: | Antibody Fragment 1B2, Heavy Chain, Light Chain, ... | | Authors: | Cogan, D.P, Soohoo, A.M, Chen, M, Brodsky, K.L, Liu, Y, Khosla, C. | | Deposit date: | 2023-08-05 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural basis for intermodular communication in assembly-line polyketide biosynthesis.
Nat.Chem.Biol., 2024
|
|
5MSB
 
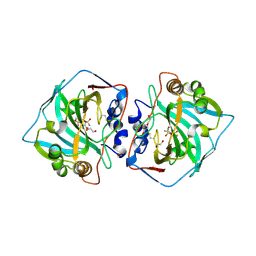 | | Crystal structure of human carbonic anhydrase isozyme XII with 2,3,5,6-tetrafluoro-4[(2-hydroxyethyl)sulfonyl]benzenesulfonamide | | Descriptor: | 1,2-ETHANEDIOL, 2,3,5,6-tetrafluoro-4-[(2-hydroxyethyl)sulfonyl]benzenesulfonamide, Carbonic anhydrase 12, ... | | Authors: | Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure correlations with the intrinsic thermodynamics of human carbonic anhydrase inhibitor binding.
PeerJ, 6, 2018
|
|
2FZJ
 
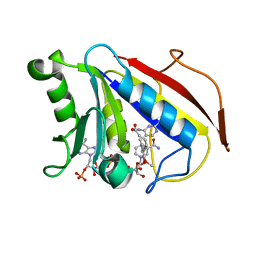 | | New Insights into DHFR Interactions: Analysis of Pneumocystis carinii and Mouse DHFR Complexes with NADPH and Two Highly Potent Trimethoprim Derivatives | | Descriptor: | 2,4-DIAMINO-5-[3',4'-DIMETHOXY-5'-(5-CARBOXYL-1-PENTYNYL)]BENZYL PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Chisum, K, Rosowsky, A. | | Deposit date: | 2006-02-09 | | Release date: | 2006-12-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | New insights into DHFR interactions: analysis of Pneumocystis carinii and mouse DHFR complexes with NADPH and two highly potent 5-(omega-carboxy(alkyloxy) trimethoprim derivatives reveals conformational correlations with activity and novel parallel ring stacking interactions.
Proteins, 65, 2006
|
|
8T2V
 
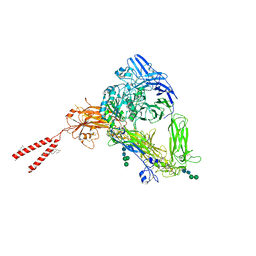 | | Cryo-EM Structures of Full-length Integrin alphaIIbbeta3 in Native Lipids | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adair, B, Xiong, J.P, Yeager, M, Arnaout, M.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of full-length integrin alpha IIb beta 3 in native lipids.
Nat Commun, 14, 2023
|
|
6W66
 
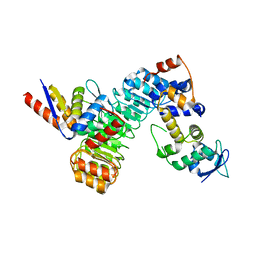 | | The structure of the F64A, S172A mutant Keap1-BTB domain in complex with SKP1-FBXL17 | | Descriptor: | F-box/LRR-repeat protein 17, Kelch-like ECH-associated protein 1, S-phase kinase-associated protein 1 | | Authors: | Mena, E.L, Gee, C.L, Kuriyan, J, Rape, M. | | Deposit date: | 2020-03-16 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Structural basis for dimerization quality control.
Nature, 586, 2020
|
|
8T2U
 
 | | Cryo-EM Structures of Full-length Integrin alphaIIbbeta3 in Native Lipids complexed with Eptifibatide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Adair, B, Xiong, J.P, Yeager, M, Arnaout, M.A. | | Deposit date: | 2023-06-06 | | Release date: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of full-length integrin alpha IIb beta 3 in native lipids.
Nat Commun, 14, 2023
|
|
6QMD
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{R})-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, DIMETHYL SULFOXIDE, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QME
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chloranyl-3-methyl-phenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, Kelch-like ECH-associated protein 1, SULFATE ION | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6O7R
 
 | | Nitrogenase MoFeP mutant F99Y, S188A from Azotobacter vinelandii in the dithionite reduced state | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Williamson, L.M, Tezcan, F.A. | | Deposit date: | 2019-03-08 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Redox-Dependent Metastability of the Nitrogenase P-Cluster.
J.Am.Chem.Soc., 141, 2019
|
|
6O7S
 
 | |
3UGJ
 
 | | Formyl Glycinamide ribonucletide amidotransferase from Salmonella Typhimurum: Role of the ATP complexation and glutaminase domain in catalytic coupling | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Morar, M, Tanwar, A.S, Panjikar, S, Anand, R. | | Deposit date: | 2011-11-02 | | Release date: | 2012-06-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Formylglycinamide ribonucleotide amidotransferase from Salmonella typhimurium: role of ATP complexation and the glutaminase domain in catalytic coupling
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6QMJ
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(7-methoxy-1-methyl-benzotriazol-5-yl)-3-[4-methyl-3-[[methyl(phenylsulfonyl)amino]methyl]phenyl]propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
1KJI
 
 | | Crystal structure of glycinamide ribonucleotide transformylase in complex with Mg-AMPPCP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CHLORIDE ION, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
1KJ9
 
 | | Crystal structure of purt-encoded glycinamide ribonucleotide transformylase complexed with Mg-ATP | | Descriptor: | 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Thoden, J.B, Firestine, S.M, Benkovic, S.J, Holden, H.M. | | Deposit date: | 2001-12-04 | | Release date: | 2002-06-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | PurT-encoded glycinamide ribonucleotide transformylase. Accommodation of adenosine nucleotide analogs within the active site.
J.Biol.Chem., 277, 2002
|
|
