5U22
 
 | | Structure of N2152 from Neocallimastix frontalis | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, IODIDE ION, ... | | Authors: | Jones, D.R, Abbott, D.W. | | Deposit date: | 2016-11-29 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.748 Å) | | Cite: | Discovery and characterization of family 39 glycoside hydrolases from rumen anaerobic fungi with polyspecific activity on rare arabinosyl substrates.
J. Biol. Chem., 292, 2017
|
|
8GQU
 
 | | AK-42 inhibitor binding human ClC-2 TMD | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-phenylmethoxy-phenyl]amino]pyridine-3-carboxylic acid, Chloride channel protein 2 | | Authors: | Wang, L. | | Deposit date: | 2022-08-30 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of ClC-2 chloride channel reveal the blocking mechanism of its specific inhibitor AK-42.
Nat Commun, 14, 2023
|
|
8AE2
 
 | | Cryo-EM structure of full-length human immunoglobulin M - F(ab')2 conformation 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, IgM C2-domain from mouse, ... | | Authors: | Chen, Q, Rosenthal, P, Tolar, P. | | Deposit date: | 2022-07-12 | | Release date: | 2022-10-26 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryomicroscopy reveals the structural basis for a flexible hinge motion in the immunoglobulin M pentamer.
Nat Commun, 13, 2022
|
|
6B5I
 
 | |
3WC8
 
 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
8DF0
 
 | | Abp1D receptor binding domain | | Descriptor: | Abp1D Receptor Binding Domain | | Authors: | Tamadonfar, K.O, Pinkner, J.S, Dodson, K.W, Hultgren, S.J. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function correlates of fibrinogen binding by Acinetobacter adhesins critical in catheter-associated urinary tract infections.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4QBO
 
 | | VRR_NUC domain | | Descriptor: | MAGNESIUM ION, Nuclease | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
1K05
 
 | |
8DKA
 
 | |
8AB2
 
 | | Crystal Structure of the Lactate Dehydrogenase of Cyanobacterium Aponinum in its apo form. | | Descriptor: | 1,2-ETHANEDIOL, L-lactate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Robin, A.Y, Girard, E, Madern, D. | | Deposit date: | 2022-07-04 | | Release date: | 2022-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Deciphering Evolutionary Trajectories of Lactate Dehydrogenases Provides New Insights into Allostery.
Mol.Biol.Evol., 40, 2023
|
|
8CWL
 
 | | Cryo-EM structure of Human 15-PGDH in complex with small molecule SW222746 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 15-hydroxyprostaglandin dehydrogenase [NAD(+)], 2-methyl-6-[7-(piperidine-1-carbonyl)quinoxalin-2-yl]isoquinolin-1(2H)-one | | Authors: | Huang, W, Taylor, D.J. | | Deposit date: | 2022-05-19 | | Release date: | 2023-03-08 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Small molecule inhibitors of 15-PGDH exploit a physiologic induced-fit closing system.
Nat Commun, 14, 2023
|
|
5TQV
 
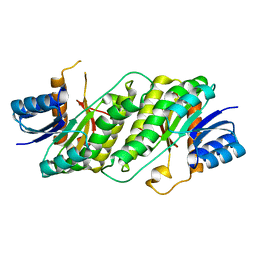 | |
8DEY
 
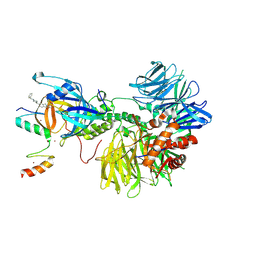 | | Ternary complex structure of Cereblon-DDB1 bound to IKZF2(ZF2,3) and the molecular glue DKY709 | | Descriptor: | (3S)-3-[5-(1-benzylpiperidin-4-yl)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Ma, X, Ornelas, E, Clifton, M.C. | | Deposit date: | 2022-06-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Discovery and characterization of a selective IKZF2 glue degrader for cancer immunotherapy.
Cell Chem Biol, 30, 2023
|
|
4QET
 
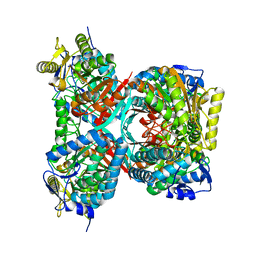 | |
8D3T
 
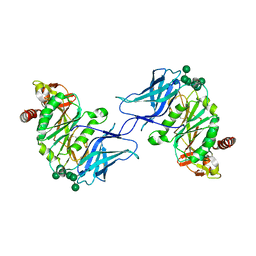 | | Crystal structure of GalS1 from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
8B2P
 
 | |
3WF7
 
 | | Crystal structure of S6K1 kinase domain in complex with a purine derivative 1-(9H-purin-6-yl)-N-[3-(trifluoromethyl)phenyl]piperidine-4-carboxamide | | Descriptor: | 1-(9H-purin-6-yl)-N-[3-(trifluoromethyl)phenyl]piperidine-4-carboxamide, GLYCEROL, Ribosomal protein S6 kinase beta-1, ... | | Authors: | Niwa, H, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2013-07-17 | | Release date: | 2014-08-06 | | Last modified: | 2014-10-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the S6K1 kinase domain in complexes with inhibitors
J.Struct.Funct.Genom., 15, 2014
|
|
8D3Z
 
 | | Crystal structure of GalS1 in complex with Manganese from Populus trichocarpas | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Galactan synthase, ... | | Authors: | Pereira, J.H, Prabhakar, P.K, Urbanowicz, B.R, Adams, P.D. | | Deposit date: | 2022-06-01 | | Release date: | 2023-03-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and biochemical insight into a modular beta-1,4-galactan synthase in plants.
Nat.Plants, 9, 2023
|
|
6B7V
 
 | | Structure of hen egg-white lysozyme pre-treated with high-pressure homogenization at 120 MPa | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Morais, M.A.B, Nascimento, A.F.Z, Tominaga, C.Y, Cristianini, M, Tribst, A.A.L, Murakami, M.T. | | Deposit date: | 2017-10-05 | | Release date: | 2018-07-25 | | Method: | X-RAY DIFFRACTION (1.482 Å) | | Cite: | How high pressure pre-treatments affect the function and structure of hen egg-white lysozyme
Innov Food Sci Emerg Technol, 47, 2018
|
|
1JN7
 
 | |
8AF6
 
 | | Room temperature SSX structure of GH11 xylanase from Nectria haematococca (4000 frames) | | Descriptor: | Endo-1,4-beta-xylanase | | Authors: | Oberthuer, D, Andaleeb, H, Betzel, C, Perbandt, M, Yefanov, O, Zielinski, K. | | Deposit date: | 2022-07-15 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rapid and efficient room-temperature serial synchrotron crystallography using the CFEL TapeDrive.
Iucrj, 9, 2022
|
|
8DJD
 
 | |
5MBC
 
 | | Structure of a bacterial light-regulated adenylyl cylcase | | Descriptor: | Beta subunit of photoactivated adenylyl cyclase, CHLORIDE ION, FLAVIN MONONUCLEOTIDE | | Authors: | Lindner, R, Hartmann, E, Tarnawski, M, Winkler, A, Frey, D, Reinstein, J, Meinhart, A, Schlichting, I. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase.
J. Mol. Biol., 429, 2017
|
|
8DJE
 
 | | CRYSTAL STRUCTURE OF GLYCOGEN SYNTHASE KINASE 3 BETA COMPLEXED WITH 3-[(CYCLOPROPYLMETHYL)AMINO] -N-(4-PHENYLPYRIDIN-3-YL)IMIDAZO[1,2-B]PYRIDAZINE-8-CARBOX AMIDE | | Descriptor: | (4S)-3-[(cyclopropylmethyl)amino]-N-(4-phenylpyridin-3-yl)imidazo[1,2-b]pyridazine-8-carboxamide, Glycogen synthase kinase-3 beta | | Authors: | Lewis, H.A, Muckelbauer, J.K. | | Deposit date: | 2022-06-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Design, Structure-Activity Relationships, and In Vivo Evaluation of Potent and Brain-Penetrant Imidazo[1,2- b ]pyridazines as Glycogen Synthase Kinase-3 beta (GSK-3 beta ) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8B27
 
 | | Dihydroprecondylocarpine acetate synthase from Catharanthus roseus | | Descriptor: | Dehydroprecondylocarpine acetate synthase, SULFATE ION | | Authors: | Langley, C, Basquin, J, Caputi, L, O'Connor, S.E. | | Deposit date: | 2022-09-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Expansion of the Catalytic Repertoire of Alcohol Dehydrogenases in Plant Metabolism.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
