4DFC
 
 | | Core UvrA/TRCF complex | | Descriptor: | Transcription-repair-coupling factor, UvrABC system protein A | | Authors: | Deaconescu, A.M, Grigorieff, N. | | Deposit date: | 2012-01-23 | | Release date: | 2012-05-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.803 Å) | | Cite: | Nucleotide excision repair (NER) machinery recruitment by the transcription-repair coupling factor involves unmasking of a conserved intramolecular interface.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5LPG
 
 | | Structure of NUDT15 in complex with 6-thio-GMP | | Descriptor: | MAGNESIUM ION, Probable 8-oxo-dGTP diphosphatase NUDT15, [(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-sulfanyl-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Masuyer, G, Carter, M, Rehling, D, Stenmark, P, Helleday, T, Jemth, A.-S, Valerie, N.C.K, Homan, E, Herr, P, Bevc, L, Page, B.D.G, Hagenkort, A. | | Deposit date: | 2016-08-12 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NUDT15 Hydrolyzes 6-Thio-DeoxyGTP to Mediate the Anticancer Efficacy of 6-Thioguanine.
Cancer Res., 76, 2016
|
|
5LUS
 
 | | Structures of DHBN domain of Pelecanus crispus BLM helicase | | Descriptor: | BLM helicase | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.433 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
6XUF
 
 | | HumRadA1 in complex with 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine in P21 | | Descriptor: | 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Scott, D.E, Hyvonen, M.E. | | Deposit date: | 2020-01-19 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.241 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To be published
|
|
4E1M
 
 | |
6XUJ
 
 | | HumRadA1 in complex with 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine in P21212 | | Descriptor: | 5-Ethyl-N-(1H-indol-5-ylmethyl)-1,3,4-thiadiazol-2-amine, DNA repair and recombination protein RadA, PHOSPHATE ION | | Authors: | Marsh, M.E, Scott, D.E, Hyvonen, M. | | Deposit date: | 2020-01-20 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Optimising crystallographic systems for structure-guided drug discovery
To be published
|
|
2KQC
 
 | | Second PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
2KQB
 
 | | First PBZ domain of human APLF protein | | Descriptor: | Aprataxin and PNK-like factor, ZINC ION | | Authors: | Neuhaus, D, Eustermann, S, Brockmann, C, Yang, J. | | Deposit date: | 2009-11-04 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose).
Nat.Struct.Mol.Biol., 17, 2010
|
|
5M89
 
 | | Spliceosome component | | Descriptor: | Spliceosome WD40 Sc | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.605 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
5M88
 
 | | Spliceosome component | | Descriptor: | Core | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
3F8T
 
 | |
3JVR
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | (1S)-1-(1H-benzimidazol-2-yl)ethyl (3,4-dichlorophenyl)carbamate, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
3SYY
 
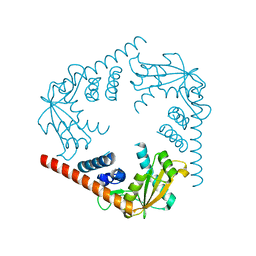 | | Crystal Structure of an alkaline exonuclease (LHK-Exo) from Laribacter hongkongensis | | Descriptor: | Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3SZ4
 
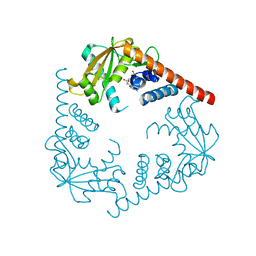 | | Crystal Structure of LHK-Exo in complex with dAMP | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, Exonuclease, MAGNESIUM ION | | Authors: | Yang, W, Chen, W.Y, Wang, H, Zhang, Q, Zhou, W, Bartlam, M, Watt, R.M, Rao, Z. | | Deposit date: | 2011-07-18 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structural and functional insight into the mechanism of an alkaline exonuclease from Laribacter hongkongensis.
Nucleic Acids Res., 39, 2011
|
|
3JCA
 
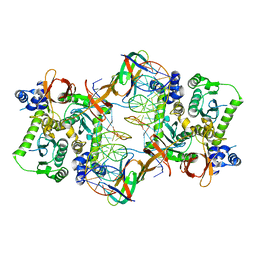 | | Core model of the Mouse Mammary Tumor Virus intasome | | Descriptor: | 5'-D(*AP*AP*TP*GP*CP*CP*GP*CP*AP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*CP*TP*G)-3', 5'-D(*CP*AP*GP*GP*TP*CP*GP*GP*CP*CP*GP*AP*CP*TP*GP*CP*GP*GP*CP*A)-3', Integrase, ... | | Authors: | Lyumkis, D.L, Ballandras-Colas, A, Brown, M, Cook, N.J, Dewdney, T.G, Demeler, B, Cherepanov, P, Engelman, A.N. | | Deposit date: | 2015-11-24 | | Release date: | 2016-02-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cryo-EM reveals a novel octameric integrase structure for betaretroviral intasome function.
Nature, 530, 2016
|
|
5J4K
 
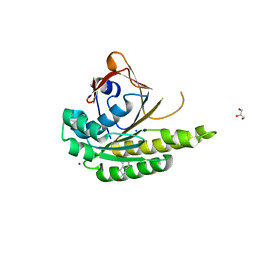 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
1UPG
 
 | |
3JVS
 
 | | Characterization of the Chk1 allosteric inhibitor binding site | | Descriptor: | 2-[(4-tert-butyl-3-nitrophenyl)carbonyl]-N-naphthalen-1-ylhydrazinecarboxamide, Serine/threonine-protein kinase Chk1 | | Authors: | Chen, P. | | Deposit date: | 2009-09-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the CHK1 allosteric inhibitor binding site.
Biochemistry, 48, 2009
|
|
5JEC
 
 | | Apo-structure of humanised RadA-mutant humRadA33F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
3K16
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
5M8C
 
 | | Spliceosome component | | Descriptor: | Pre-mRNA-processing factor 19 | | Authors: | Moura, T.R, Pena, V. | | Deposit date: | 2016-10-28 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Prp19/Pso4 Is an Autoinhibited Ubiquitin Ligase Activated by Stepwise Assembly of Three Splicing Factors.
Mol. Cell, 69, 2018
|
|
4E1N
 
 | |
1US6
 
 | |
2IO5
 
 | | Crystal structure of the CIA- histone H3-H4 complex | | Descriptor: | ASF1A protein, Histone H3.1, Histone H4 | | Authors: | Natsume, R, Akai, Y, Horikoshi, M, Senda, T. | | Deposit date: | 2006-10-10 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4.
Nature, 446, 2007
|
|
