3NXR
 
 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NXX
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro-2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)-4-methylpent-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1A0M
 
 | | 1.1 ANGSTROM CRYSTAL STRUCTURE OF A-CONOTOXIN [TYR15]-EPI | | Descriptor: | ALPHA-CONOTOXIN [TYR15]-EPI | | Authors: | Hu, S.-H, Loughnan, M, Miller, R, Weeks, C.M, Blessing, R.H, Alewood, P.F, Lewis, R.J, Martin, J.L. | | Deposit date: | 1997-12-03 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The 1.1 A resolution crystal structure of [Tyr15]EpI, a novel alpha-conotoxin from Conus episcopatus, solved by direct methods.
Biochemistry, 37, 1998
|
|
5OVB
 
 | | Crystal structure of human BRD4(1) bromodomain in complex with DR46 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-[3-(5-ethanoyl-2-ethoxy-phenyl)-5-(1-methylpyrazol-3-yl)phenyl]furan-2-carboxamide | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2017-08-28 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Chemical Space Expansion of Bromodomain Ligands Guided by in Silico Virtual Couplings (AutoCouple).
Acs Cent.Sci., 4, 2018
|
|
4GKH
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, with substrate kanamycin and small molecule inhibitor 1-NA-PP1 | | Descriptor: | 1-tert-butyl-3-(naphthalen-1-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, ACETATE ION, Aminoglycoside 3'-phosphotransferase AphA1-IAB, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Minasov, G, Egorova, O, Di Leo, R, Shakya, T, Spanogiannopoulos, P, Todorovic, N, Capretta, A, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-11 | | Release date: | 2012-09-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.863 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
6JFJ
 
 | | Crystal structure of Pullulanase from Paenibacillus barengoltzii complex with maltohexaose and alpha-cyclodextrin | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wu, S.W, Yang, S.Q, Qin, Z, You, X, Huang, P, Jiang, Z.Q. | | Deposit date: | 2019-02-09 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Structural basis of carbohydrate binding in domain C of a type I pullulanase from Paenibacillus barengoltzii.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
3GMD
 
 | | Structure-based design of 7-Azaindole-pyrrolidines as inhibitors of 11beta-Hydroxysteroid-Dehydrogenase type I | | Descriptor: | 2-methyl-3-{(3S)-1-[(1-pyridin-2-ylcyclopropyl)carbonyl]pyrrolidin-3-yl}-1H-pyrrolo[2,3-b]pyridine, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Valeur, E, Lepifre, F, Roche, D, Christmann-Franck, S, Hillertz, P, Musil, D. | | Deposit date: | 2009-03-13 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure-based design of 7-azaindole-pyrrolidine amides as inhibitors of 11 beta-hydroxysteroid dehydrogenase type I.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4M10
 
 | | Crystal Structure of Murine Cyclooxygenase-2 Complex with Isoxicam | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-hydroxy-2-methyl-N-(5-methyl-1,2-oxazol-3-yl)-2H-1,2-benzothiazine-3-carboxamide 1,1-dioxide, ... | | Authors: | Xu, S, Hermanson, D.J, Banerjee, S, Ghebreelasie, K, Marnett, L.J. | | Deposit date: | 2013-08-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Oxicams Bind in a Novel Mode to the Cyclooxygenase Active Site via a Two-water-mediated H-bonding Network.
J.Biol.Chem., 289, 2014
|
|
3NVV
 
 | | Crystal Structure of Bovine Xanthine Oxidase in Complex with Arsenite | | Descriptor: | ARSENITE, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Cao, H, Hille, R. | | Deposit date: | 2010-07-08 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | X-ray Crystal Structure of Arsenite-Inhibited Xanthine Oxidase: Mu-Sulfido,Mu-Oxo Double Bridge between Molybdenum and Arsenic in the Active Site.
J.Am.Chem.Soc., 133, 2011
|
|
2C3C
 
 | | 2.01 Angstrom X-ray crystal structure of a mixed disulfide between coenzyme M and NADPH-dependent oxidoreductase 2-ketopropyl coenzyme M carboxylase | | Descriptor: | 1-THIOETHANESULFONIC ACID, 2-OXOPROPYL-COM REDUCTASE, ACETONE, ... | | Authors: | Pandey, A.S, Nocek, B, Clark, D.D, Ensign, S.A, Peters, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-12-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic Implications of the Structure of the Mixed-Disulfide Intermediate of the Disulfide Oxidoreductase, 2-Ketopropyl-Coenzyme M Oxidoreductase/Carboxylase.
Biochemistry, 45, 2006
|
|
4GLO
 
 | | Crystal structure of a short chain dehydrogenase homolog (target EFI-505321) from burkholderia multivorans, with bound NAD | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier protein] reductase, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Hobbs, M.E, Morisco, L.L, Wasserman, S.R, Sojitra, S, Imker, H.J, Raushel, F.M, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-08-14 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a short chain dehydrogenase homolog
(target EFI-505321) from burkholderia multivorans, with bound NAD
To be Published
|
|
3HEK
 
 | |
5GR1
 
 | |
3NXO
 
 | | Perferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1Z)-2-(2-methoxyphenyl)-3-methylbut-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3NXV
 
 | | Preferential Selection of Isomer Binding from Chiral Mixtures: Alternate Binding Modes Observed for the E- and Z-isomers of a Series of 5-Substituted 2,4-Diaminofuro[2,3-d]pyrimidines as Ternary Complexes with NADPH and Human Dihydrofolate Reductase | | Descriptor: | 5-[(1E)-2-(2-methoxyphenyl)hex-1-en-1-yl]furo[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-14 | | Release date: | 2010-12-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Preferential selection of isomer binding from chiral mixtures: alternate binding modes observed for the E and Z isomers of a series of 5-substituted 2,4-diaminofuro[2,3-d]pyrimidines as ternary complexes with NADPH and human dihydrofolate reductase.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
4EKS
 
 | | T4 Lysozyme L99A/M102H with Isoxazole Bound | | Descriptor: | 1,2-benzisoxazole, 2-HYDROXYETHYL DISULFIDE, ACETATE ION, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Engineering a model protein cavity to catalyze the Kemp elimination.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3O5A
 
 | | Crystal Structure of partially reduced Periplasmic Nitrate Reductase from Cupriavidus necator using Ionic Liquids | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CHLORIDE ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-07-28 | | Release date: | 2011-04-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
4GUH
 
 | | 1.95 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) E86A Mutant in Complex with Dehydroshikimate (Crystal Form #2) | | Descriptor: | (4S,5R)-4,5-dihydroxy-3-oxocyclohex-1-ene-1-carboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Reassessing the type I dehydroquinate dehydratase catalytic triad: Kinetic and structural studies of Glu86 mutants.
Protein Sci., 22, 2013
|
|
4GUI
 
 | | 1.78 Angstrom Crystal Structure of the Salmonella enterica 3-Dehydroquinate Dehydratase (aroD) in Complex with Quinate | | Descriptor: | (1S,3R,4S,5R)-1,3,4,5-tetrahydroxycyclohexanecarboxylic acid, 3-dehydroquinate dehydratase, NICKEL (II) ION | | Authors: | Light, S.H, Minasov, G, Duban, M.-E, Shuvalova, L, Kwon, K, Lavie, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-29 | | Release date: | 2012-09-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structures of type I dehydroquinate dehydratase in complex with quinate and shikimate suggest a novel mechanism of schiff base formation.
Biochemistry, 53, 2014
|
|
5UAS
 
 | | Structure of a new family of Polysaccharide lyase PL25-Ulvanlyase bound to -[GlcA(1-4)Rha3S]- | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CHLORIDE ION, ... | | Authors: | Ulaganathan, T.S, Cygler, M. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Ulvan-Degrading Polysaccharide Lyase Family: Structure and Catalytic Mechanism Suggests Convergent Evolution of Active Site Architecture.
ACS Chem. Biol., 12, 2017
|
|
1FDR
 
 | | FLAVODOXIN REDUCTASE FROM E. COLI | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVODOXIN REDUCTASE | | Authors: | Ingelman, M, Bianchi, V, Eklund, H. | | Deposit date: | 1997-03-06 | | Release date: | 1997-09-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The three-dimensional structure of flavodoxin reductase from Escherichia coli at 1.7 A resolution.
J.Mol.Biol., 268, 1997
|
|
5QIH
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of HAO1 in complex with Z2697514548 | | Descriptor: | 1-methylindazole-3-carboxamide, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | MacKinnon, S, Bezerra, G.A, Krojer, T, Bradley, A.R, Talon, R, Brandao-Neto, J, Douangamath, A, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Oppermann, U, Brennan, P.E, Yue, W.W. | | Deposit date: | 2018-05-22 | | Release date: | 2018-07-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
6EN5
 
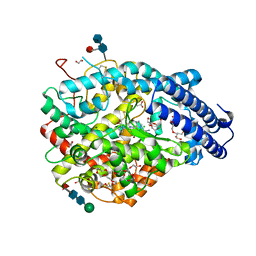 | | Crystal structure A of the Angiotensin-1 converting enzyme N-domain in complex with a diprolyl inhibitor. | | Descriptor: | (2~{S})-1-[(2~{S})-2-[[(1~{S})-1-[(2~{S})-1-[(2~{S})-2-azanyl-4-oxidanyl-4-oxidanylidene-butanoyl]pyrrolidin-2-yl]-2-oxidanyl-2-oxidanylidene-ethyl]amino]propanoyl]pyrrolidine-2-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Cozier, G.E, Acharya, K.R, Fienberg, S, Chibale, K, Sturrock, E.D. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Design and Development of a Potent and Selective Novel Diprolyl Derivative That Binds to the N-Domain of Angiotensin-I Converting Enzyme.
J. Med. Chem., 61, 2018
|
|
1K5P
 
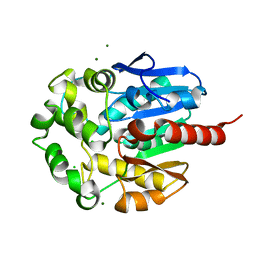 | | Hydrolytic haloalkane dehalogenase LINB from sphingomonas paucimobilis UT26 at 1.8A resolution | | Descriptor: | 1,3,4,6-tetrachloro-1,4-cyclohexadiene hydrolase, CHLORIDE ION, MAGNESIUM ION | | Authors: | Streltsov, V.A, Damborsky, J, Wilce, M.C.J. | | Deposit date: | 2001-10-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Haloalkane dehalogenase LinB from Sphingomonas paucimobilis UT26: X-ray crystallographic studies of dehalogenation of brominated substrates
Biochemistry, 42, 2003
|
|
6KIF
 
 | | Structure of cyanobacterial photosystem I-IsiA-flavodoxin supercomplex | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Cao, P, Cao, D.F, Si, L, Su, X.D, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2019-07-18 | | Release date: | 2020-02-12 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for energy and electron transfer of the photosystem I-IsiA-flavodoxin supercomplex.
Nat.Plants, 6, 2020
|
|
