1NOA
 
 | |
1NZ2
 
 | | K45E Variant of Horse Heart Myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, M, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1O9G
 
 | |
1QX4
 
 | | Structrue of S127P mutant of cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase | | Authors: | Bewley, M.C, Davis, C.A, Marohnic, C.C, Taormina, D, Barber, M.J. | | Deposit date: | 2003-09-04 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the S127P mutant of cytochrome b5 reductase that causes methemoglobinemia shows the AMP moiety of the flavin occupying the substrate binding site
Biochemistry, 42, 2003
|
|
1QTY
 
 | |
1QY2
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOPROPYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1R0Q
 
 | | Characterization of the conversion of the malformed, recombinant cytochrome rc552 to a 2-formyl-4-vinyl (Spirographis) heme | | Descriptor: | 2-FORMYL-PROTOPORPHRYN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-22 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QY1
 
 | | Thermodynamics of Binding of 2-methoxy-3-isopropylpyrazine and 2-methoxy-3-isobutylpyrazine to the Major Urinary Protein | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, Major Urinary Protein, ... | | Authors: | Bingham, R.J, Findlay, J.B.C, Hsieh, S.-Y, Kalverda, A.P, Kjellberg, A, Perazzolo, C, Phillips, S.E.V, Seshadri, K, Trinh, C.H, Turnbull, W.B, Bodenhausen, G, Homans, S.W. | | Deposit date: | 2003-09-09 | | Release date: | 2004-02-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Thermodynamics of Binding of 2-Methoxy-3-isopropylpyrazine and 2-Methoxy-3-isobutylpyrazine to the Major Urinary Protein.
J.Am.Chem.Soc., 126, 2004
|
|
1QX5
 
 | |
1QYZ
 
 | | Characterization of the malformed, recombinant cytochrome rC552 | | Descriptor: | 2-ACETYL-PROTOPORPHYRIN IX, Cytochrome c-552 | | Authors: | Fee, J.A, Todaro, T.R, Luna, E, Sanders, D, Hunsicker-Wang, L.M, Patel, K.M, Bren, K.L, Gomez-Moran, E, Hill, M.G, Ai, J, Loehr, T.M, Oertling, W.A, Williams, P.A, Stout, C.D, McRee, D, Pastuszyn, A. | | Deposit date: | 2003-09-12 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cytochrome rC552, formed during expression of the truncated, Thermus thermophilus cytochrome c552 gene in the cytoplasm of Escherichia coli, reacts spontaneously to form protein-bound 2-formyl-4-vinyl (Spirographis) heme.
Biochemistry, 43, 2004
|
|
1QLQ
 
 | | Bovine Pancreatic Trypsin Inhibitor (BPTI) Mutant with Altered Binding Loop Sequence | | Descriptor: | PANCREATIC TRYPSIN INHIBITOR, SULFATE ION | | Authors: | Czapinska, H, Krzywda, S, Sheldrick, G.M, Otlewski, J, Jaskolski, M. | | Deposit date: | 1999-09-10 | | Release date: | 1999-10-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High Resolution Structure of Bovine Pancreatic Trypsin Inhibitor with Altered Binding Loop Sequence
J.Mol.Biol., 295, 1999
|
|
1QLS
 
 | | S100C (S100A11),OR CALGIZZARIN, IN COMPLEX WITH ANNEXIN I N-TERMINUS | | Descriptor: | ANNEXIN I, CALCIUM ION, S100C PROTEIN | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1999-09-15 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Ca2+ Dependent Association between S100C (S100A11) and its Target, the N-Terminal Part of Annexin I
Structure, 8, 2000
|
|
2CK2
 
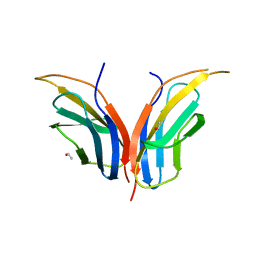 | | Structure of core-swapped mutant of fibronectin | | Descriptor: | ACETYL GROUP, HUMAN FIBRONECTIN | | Authors: | Ng, S.P, Billings, K.S, Ohashi, T, Allen, M.D, Best, R.B, Randles, L.G, Erickson, H.P, Clarke, J. | | Deposit date: | 2006-04-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing an Extracellular Matrix Protein with Enhanced Mechanical Stability
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
4GM1
 
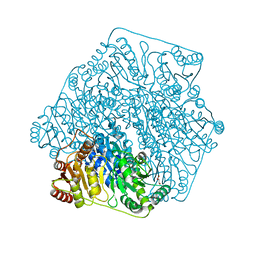 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403S | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
4GG1
 
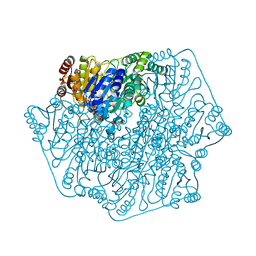 | | Crystal Structure of Benzoylformate Decarboxylase Mutant L403T | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, Benzoylformate decarboxylase, CALCIUM ION, ... | | Authors: | Novak, W.R.P, Andrews, F.H, Tom, A.R, Gunderman, P.R, McLeish, M.J. | | Deposit date: | 2012-08-04 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | A bulky hydrophobic residue is not required to maintain the v-conformation of enzyme-bound thiamin diphosphate.
Biochemistry, 52, 2013
|
|
101D
 
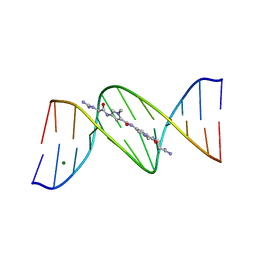 | |
136D
 
 | |
144D
 
 | | MINOR GROOVE BINDING OF SN6999 TO AN ALKYLATED DNA: MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-SN6999 COMPLEX | | Descriptor: | 1-METHYL-4-[4-[4-(4-(1-METHYLQUINOLINIUM)AMINO)BENZAMIDO]ANILINO]PYRIDINIUM, DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Gao, Y.-G, Sriram, M, Denny, W.A, Wang, A.H.-J. | | Deposit date: | 1993-10-26 | | Release date: | 1995-05-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Minor groove binding of SN6999 to an alkylated DNA: molecular structure of d(CGC[e6G]AATTCGCG)-SN6999 complex.
Biochemistry, 32, 1993
|
|
190D
 
 | |
186D
 
 | |
1C2X
 
 | |
1BMB
 
 | | GRB2-SH2 DOMAIN IN COMPLEX WITH KPFY*VNVEF (PKF270-974) | | Descriptor: | PROTEIN (GROWTH FACTOR RECEPTOR BOUND PROTEIN 2), PROTEIN (PKF270-974) | | Authors: | Rondeau, J.M, Zurini, M. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and conformational requirements for high-affinity binding to the SH2 domain of Grb2(1).
J.Med.Chem., 42, 1999
|
|
1BM2
 
 | |
1A5P
 
 | | C[40,95]A VARIANT OF BOVINE PANCREATIC RIBONUCLEASE A | | Descriptor: | RIBONUCLEASE A, SULFATE ION | | Authors: | Pearson, M.A, Karplus, P.A, Dodge, R.W, Laity, J.H, Scheraga, H.A. | | Deposit date: | 1998-02-17 | | Release date: | 1998-05-27 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of two mutants that have implications for the folding of bovine pancreatic ribonuclease A.
Protein Sci., 7, 1998
|
|
1AML
 
 | |
