8V6T
 
 | | Crystal structure of EcThsB2 | | Descriptor: | BROMIDE ION, Molecular chaperone Tir | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural characterisation of macro domain-containing Thoeris antiphage defence systems
To Be Published
|
|
8V6S
 
 | | Crystal structure of PcThsA in complex with Imidazole Adenine Dinucleotide | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-imidazol-1-yl-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterisation of macro domain-containing Thoeris antiphage defence systems
To Be Published
|
|
8V6R
 
 | | Crystal structure of EcThsA in complex with ADPR | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, E, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structural characterisation of macro domain containing Thoeris antiphage defence systems
To Be Published
|
|
8V6Q
 
 | | Crystal structure of EcThsA in ligand-free state | | Descriptor: | Thoeris protein ThsA Macro domain-containing protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Goulart, C.C, Hartley-Tassell, L, Sorbello, M, Vasquez, V, Mishra, B.P, Holt, S, Gu, W, Kobe, B, Ve, T. | | Deposit date: | 2023-12-02 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Structural characterisation of macro domain containing Thoeris antiphage defence systems
To Be Published
|
|
8V6P
 
 | | Proteus vulgaris tryptophan indole-lyase complexed with 7-aza-L-tryptophan | | Descriptor: | (2E)-2-{[(Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methyl]imino}-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)propanoic acid, (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-3-(1H-pyrrolo[2,3-b]pyridin-3-yl)-L-alanine, DIMETHYL SULFOXIDE, ... | | Authors: | Phillips, R.S. | | Deposit date: | 2023-12-02 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Proteus vulgaris tryptophan indole-lyase complexed with L-alanine
To Be Published
|
|
8V6O
 
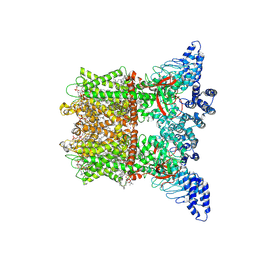 | | Inactivated-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-aminoethyl diphenylborinate, SODIUM ION, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6N
 
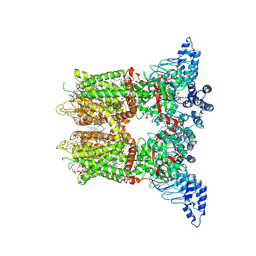 | | Open-state cryo-EM structure of human TRPV3 in presence of 2-APB in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-aminoethyl diphenylborinate, ARACHIDONIC ACID, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6M
 
 | | Inactivated-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Tetrahydrocannabivarin, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6L
 
 | | Open-state cryo-EM structure of human TRPV3 in presence of tetrahydrocannabivarin (THCV) in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Tetrahydrocannabivarin, ... | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6K
 
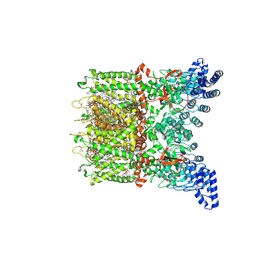 | | Apo-state cryo-EM structure of human TRPV3 in cNW30 nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, SODIUM ION, Transient receptor potential cation channel subfamily V member 3 | | Authors: | Nadezhdin, K.D, Neuberger, A, Sobolevsky, A.I. | | Deposit date: | 2023-12-01 | | Release date: | 2024-04-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | TRPV3 activation by different agonists accompanied by lipid dissociation from the vanilloid site.
Sci Adv, 10, 2024
|
|
8V6J
 
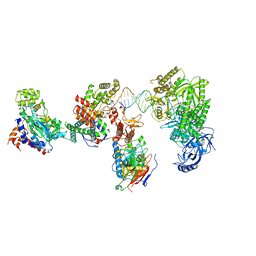 | | DNA elongation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6I
 
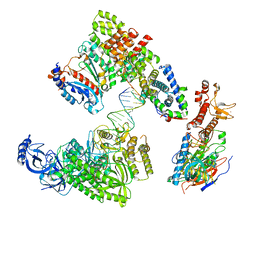 | | DNA elongation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (14.06 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6H
 
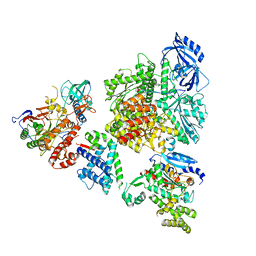 | | DNA initiation complex (configuration 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.11 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V6G
 
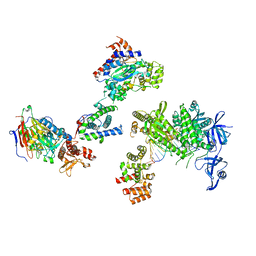 | | DNA initiation complex (configuration 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, ... | | Authors: | Mullins, E.A, Durie, C.L, Ohi, M.D, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-12-01 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (11.16 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V62
 
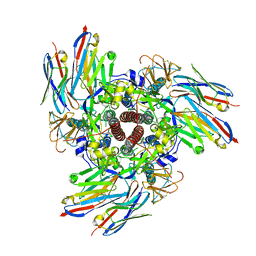 | |
8V61
 
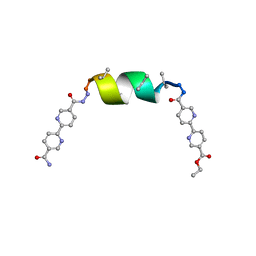 | | UIC-1 mutant - UIC-1-L6I | | Descriptor: | UIC-1-L6I | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5Z
 
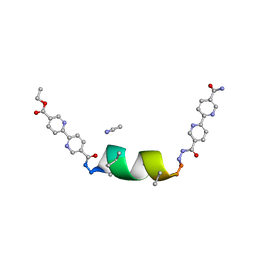 | | UIC-1 mutant - UIC-1-L6M | | Descriptor: | ACETONITRILE, UIC-1-L6M | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5Y
 
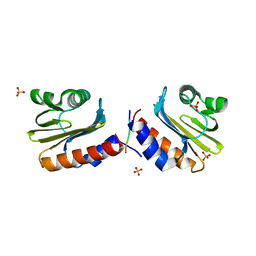 | |
8V5X
 
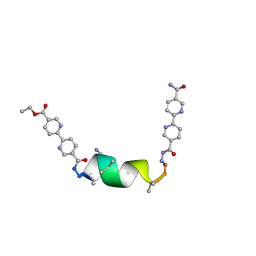 | | UIC-1 mutant - UIC-1-L6A | | Descriptor: | ACETONITRILE, UIC-1-L6A | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5W
 
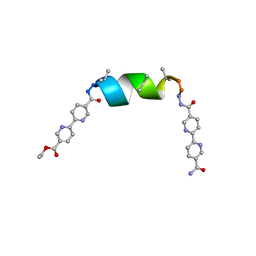 | | UIC-1 mutant UIC-1-B5T | | Descriptor: | UIC-1-B5T | | Authors: | Heinz-Kunert, S.L. | | Deposit date: | 2023-12-01 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Pore Restructuring of Peptide Frameworks by Mutations at Distal Packing Residues.
Biomacromolecules, 25, 2024
|
|
8V5O
 
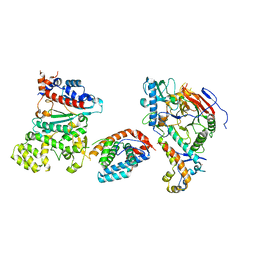 | | Tetramer core subcomplex (conformation 3) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.99 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5N
 
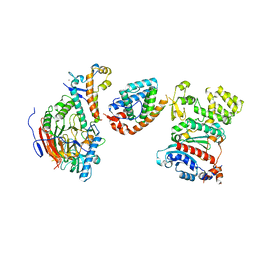 | | Tetramer core subcomplex (conformation 2) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (8.56 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5M
 
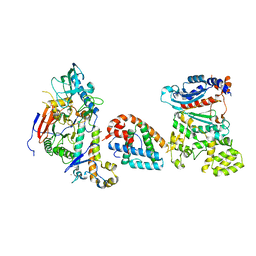 | | Tetramer core subcomplex (conformation 1) of Xenopus laevis DNA polymerase alpha-primase | | Descriptor: | DNA polymerase alpha catalytic subunit, DNA polymerase alpha subunit B, DNA primase, ... | | Authors: | Mullins, E.A, Chazin, W.J, Eichman, B.F. | | Deposit date: | 2023-11-30 | | Release date: | 2023-12-20 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (9.22 Å) | | Cite: | A mechanistic model of primer synthesis from catalytic structures of DNA polymerase alpha-primase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8V5K
 
 | |
8V5G
 
 | |
