6YNF
 
 | | GAPDH purified from the supernatant of HEK293F cells: crystal form 3 of 4. | | Descriptor: | 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Roversi, P, Lia, A. | | Deposit date: | 2020-04-13 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Partial catalytic Cys oxidation of human GAPDH to Cys-sulfonic acid.
Wellcome Open Res, 5, 2020
|
|
6B1Y
 
 | | Crystal structure KPC-2 beta-lactamase complexed with WCK 5153 by co-crystallization | | Descriptor: | (2S,5Z)-1-formyl-5-imino-N'-[(3R)-1-(sulfooxy)pyrrolidine-3-carbonyl]piperidine-2-carbohydrazide, CHLORIDE ION, Carbapenem-hydrolyzing beta-lactamase KPC, ... | | Authors: | van den Akker, F, Nguyen, N.Q. | | Deposit date: | 2017-09-19 | | Release date: | 2018-08-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Strategic Approaches to Overcome Resistance against Gram-Negative Pathogens Using beta-Lactamase Inhibitors and beta-Lactam Enhancers: Activity of Three Novel Diazabicyclooctanes WCK 5153, Zidebactam (WCK 5107), and WCK 4234.
J. Med. Chem., 61, 2018
|
|
5UI3
 
 | |
7KK3
 
 | | Structure of the catalytic domain of PARP1 in complex with talazoparib | | Descriptor: | (8S,9R)-5-fluoro-8-(4-fluorophenyl)-9-(1-methyl-1H-1,2,4-triazol-5-yl)-2,7,8,9-tetrahydro-3H-pyrido[4,3,2-de]phthalazin-3-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Gajiwala, K.S, Ryan, K. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Dissecting the molecular determinants of clinical PARP1 inhibitor selectivity for tankyrase1.
J.Biol.Chem., 296, 2021
|
|
6YO3
 
 | | LecA from Pseudomonas aeruginosa in complex with a catechol CAS no. 67984-81-0 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-bis(oxidanyl)benzenecarbonitrile, CALCIUM ION, ... | | Authors: | Kuhaudomlarp, S, Imberty, A, Titz, A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Non-Carbohydrate Glycomimetics as Inhibitors of Calcium(II)-Binding Lectins.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8RD1
 
 | | HUWE1 WWE domain in complex with compound 4 | | Descriptor: | 2-(2-hydroxy-2-oxoethyl)-1,3-bis(oxidanylidene)isoindole-5-carboxylic acid, CHLORIDE ION, E3 ubiquitin-protein ligase HUWE1 | | Authors: | Muenzker, L, Zak, K.M, Boettcher, J. | | Deposit date: | 2023-12-07 | | Release date: | 2024-07-31 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.896 Å) | | Cite: | A ligand discovery toolbox for the WWE domain family of human E3 ligases.
Commun Biol, 7, 2024
|
|
6B24
 
 | | Crystal structure of fluoride channel Fluc Ec2 F80Y Mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, Fluoride ion transporter CrcB, ... | | Authors: | Last, N.B, Sun, S, Pham, M.C, Miller, C. | | Deposit date: | 2017-09-19 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular determinants of permeation in a fluoride-specific ion channel.
Elife, 6, 2017
|
|
7KQJ
 
 | | CRYSTAL STRUCTURE OF RAR-RELATED ORPHAN RECEPTOR C (NHIS-RORGT(244-487)-L6-SRC1(678-692) IN COMPLEX WITH A NOVEL TRICYCLIC-CARBOCYLIC RORGT INVERSE AGONIST | | Descriptor: | N-[(3R,3aS,9bS)-9b-[(4-fluorophenyl)sulfonyl]-7-(1,1,1,2,3,3,3-heptafluoropropan-2-yl)-2,3,3a,4,5,9b-hexahydro-1H-cyclopenta[a]naphthalen-3-yl]-2-hydroxy-2-methylpropanamide, Nuclear receptor ROR-gamma | | Authors: | Sack, J.S. | | Deposit date: | 2020-11-16 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Tricyclic-Carbocyclic ROR gamma t Inverse Agonists-Discovery of BMS-986313.
J.Med.Chem., 64, 2021
|
|
7PUP
 
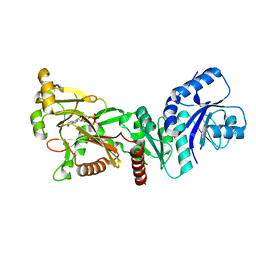 | | INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 from Arabidopsis thaliana (AtITPK4) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol 1,3,4-trisphosphate 5/6-kinase 4 | | Authors: | Whitfield, H, He, S, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversification in the inositol tris/tetrakisphosphate kinase (ITPK) family: crystal structure and enzymology of the outlier AtITPK4.
Biochem.J., 480, 2023
|
|
6B2H
 
 | |
6PJ4
 
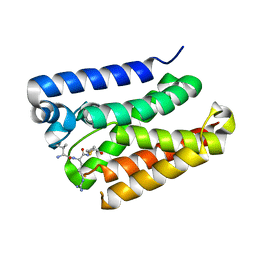 | |
7ZPH
 
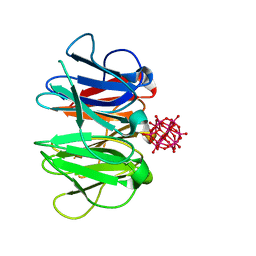 | | Crystal structure of Pizza6-TNK-RSH with Silicotungstic Acid (STA) polyoxometalate | | Descriptor: | Keggin (STA), Pizza6-TNK-RSH | | Authors: | Wouters, S.M.L, Kamata, K, Takahashi, K, Vandebroek, L, Parac-Vogt, T.N, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2022-04-27 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Mutational study of a symmetry matched protein-polyoxometalate interface
To be published
|
|
5FVW
 
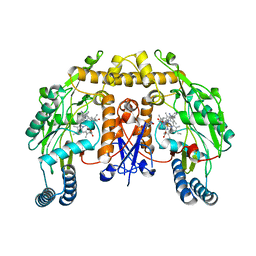 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 4-methyl-6-(2-(5-(3-(methylamino)propyl)pyridin-3-yl)ethyl) pyridin-2-amine | | Descriptor: | 4-METHYL-6-(2-(5-(3-(METHYLAMINO)PROPYL)PYRIDIN-3-YL)ETHYL)PYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, NITRIC OXIDE SYNTHASE, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
6EWR
 
 | | Putative sugar aminotransferase Spr1654 from Streptococcus pneumoniae, PMP-form | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Putative capsular polysaccharide biosynthesis protein | | Authors: | Achour, A, Sun, R, Sandalova, T, Han, X. | | Deposit date: | 2017-11-06 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional studies of Spr1654: an essential aminotransferase in teichoic acid biosynthesis inStreptococcus pneumoniae.
Open Biol, 8, 2018
|
|
7KHH
 
 | | Ternary complex of VHL/BRD4-BD1/Compound9 (4-(3,5-difluoropyridin-2-yl)-N-(11-(((S)-1-((2S,4R)-4-hydroxy-2-((4-(4-methylthiazol-5-yl)benzyl)carbamoyl)pyrrolidin-1-yl)-3,3-dimethyl-1-oxobutan-2-yl)amino)-11-oxoundecyl)-10-methyl-7-((methylsulfonyl)methyl)-11-oxo-3,4,10,11-tetrahydro-1H-1,4,10-triazadibenzo[cd,f]azulene-6-carboxamide) | | Descriptor: | Bromodomain-containing protein 4, Elongin-B, Elongin-C, ... | | Authors: | Murray, J.M. | | Deposit date: | 2020-10-21 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Antibody-Mediated Delivery of Chimeric BRD4 Degraders. Part 2: Improvement of In Vitro Antiproliferation Activity and In Vivo Antitumor Efficacy.
J.Med.Chem., 64, 2021
|
|
6BG2
 
 | |
7ZPS
 
 | |
5G57
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-001 | | Descriptor: | (4~{a}~{S},8~{a}~{R})-2-cycloheptyl-4-[4-methoxy-3-[4-[4-(1~{H}-1,2,3,4-tetrazol-5-yl)phenoxy]butoxy]phenyl]-4~{a},5,8,8~{a}-tetrahydrophthalazin-1-one, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-05-22 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
6YPE
 
 | |
6FBA
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
7ZW4
 
 | | Crystal structure of Talin R7R8 domains with Caskin-2 LD-peptide | | Descriptor: | Caskin-2, Talin-1 | | Authors: | Celie, P.H.N, Joosten, R.P, Sonnenberg, A, Perrakis, A. | | Deposit date: | 2022-05-18 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Caskin2 is a novel talin- and Abi1-binding protein that promotes cell motility.
J.Cell.Sci., 137, 2024
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
6XZN
 
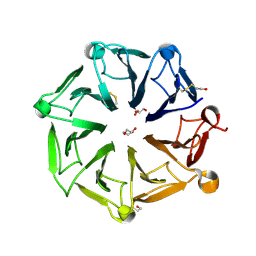 | | Arabidopsis UV-B photoreceptor UVR8 mutant G101S W285A | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, TRIETHYLENE GLYCOL, ... | | Authors: | Lau, K, Hothorn, M. | | Deposit date: | 2020-02-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A constitutively monomeric UVR8 photoreceptor confers enhanced UV-B photomorphogenesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6XZX
 
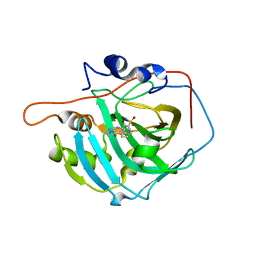 | | crystal structure of human carbonic anhydrase I in complex with 4-(3-(2-((2-fluorobenzyl)amino)ethyl)ureido) benzenesulfonamide | | Descriptor: | 1-[2-[(phenylmethyl)amino]ethyl]-3-(3-sulfamoylphenyl)urea, Carbonic anhydrase 1, ZINC ION | | Authors: | Zanotti, G, Majid, A, Bozdag, M, Angeli, A, Carta, F, Berto, P, Supuran, C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Benzylaminoethyureido-Tailed Benzenesulfonamides: Design, Synthesis, Kinetic and X-ray Investigations on Human Carbonic Anhydrases.
Int J Mol Sci, 21, 2020
|
|
6EXU
 
 | |
