3K27
 
 | | Complex structure of EED and trimethylated H3K9 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Bian, C.B, Xu, C, Qiu, W, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3JRB
 
 | |
3JRH
 
 | |
3K0A
 
 | | Crystal structure of the phosphorylation-site mutant S431A of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
3K0C
 
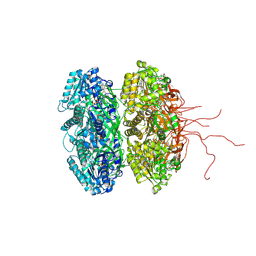 | | Crystal structure of the phosphorylation-site double mutant S431A/T432E of the KaiC circadian clock protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Circadian clock protein kinase KaiC, MAGNESIUM ION | | Authors: | Pattanayek, R, Egli, M, Pattanayek, S. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of KaiC Circadian Clock Mutant Proteins: A New Phosphorylation Site at T426 and Mechanisms of Kinase, ATPase and Phosphatase.
Plos One, 4, 2009
|
|
5OBM
 
 | | Crystal structure of Gentamicin bound to the yeast 80S ribosome | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Prokhorova, I, Djumagulov, M, Urzhumtsev, A, Yusupov, M, Yusupova, G. | | Deposit date: | 2017-06-28 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Aminoglycoside interactions and impacts on the eukaryotic ribosome.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4V9O
 
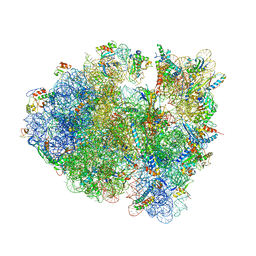 | |
4MLP
 
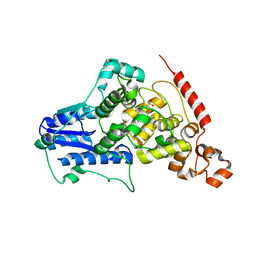 | |
4V5B
 
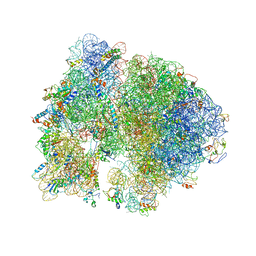 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
4WOI
 
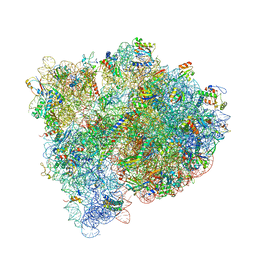 | | 4,5-linked aminoglycoside antibiotics regulate the bacterial ribosome by targeting dynamic conformational processes within intersubunit bridge B2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Pulk, A, Cate, J.H.D, Blanchard, S, Wasserman, M, Altman, R, Zhou, Z, Zinder, J, Green, K, Garneau-Tsodikova, S. | | Deposit date: | 2014-10-15 | | Release date: | 2015-08-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chemically related 4,5-linked aminoglycoside antibiotics drive subunit rotation in opposite directions.
Nat Commun, 6, 2015
|
|
4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V55
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin and ribosome recycling factor (RRF). | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-17 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V53
 
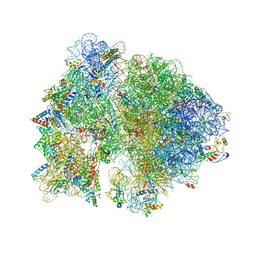 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with gentamicin. | | Descriptor: | (2R,3R,4R,5R)-2-((1S,2S,3R,4S,6R)-4,6-DIAMINO-3-((2R,3R,6S)-3-AMINO-6-(AMINOMETHYL)-TETRAHYDRO-2H-PYRAN-2-YLOXY)-2-HYDR OXYCYCLOHEXYLOXY)-5-METHYL-4-(METHYLAMINO)-TETRAHYDRO-2H-PYRAN-3,5-DIOL, 16S rRNA, 23S rRNA, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V7K
 
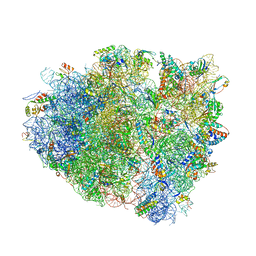 | | Structure of RelE nuclease bound to the 70S ribosome (postcleavage state) | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Neubauer, C, Gao, Y.-G, Andersen, K.R, Dunham, C.M, Kelley, A.C, Hentschel, J, Gerdes, K, Ramakrishnan, V, Brodersen, D.E. | | Deposit date: | 2009-11-02 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE.
Cell(Cambridge,Mass.), 139, 2009
|
|
8C60
 
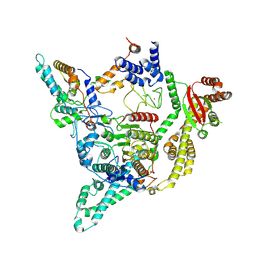 | | Cryo-EM structure of the human SIN3B full-length complex at 3.4 Angstrom resolution | | Descriptor: | CALCIUM ION, Histone deacetylase 2, Isoform 2 of Paired amphipathic helix protein Sin3b, ... | | Authors: | Alfieri, C, Wan, S.M, Muhammad, R. | | Deposit date: | 2023-01-10 | | Release date: | 2023-05-10 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mechanism of assembly, activation and lysine selection by the SIN3B histone deacetylase complex.
Nat Commun, 14, 2023
|
|
4UY7
 
 | |
2P2T
 
 | | Crystal structure of dynein light chain LC8 bound to residues 123-138 of intermediate chain IC74 | | Descriptor: | ACETATE ION, Dynein intermediate chain peptide, Dynein light chain 1, ... | | Authors: | Benison, G, Karplus, P.A, Barbar, E. | | Deposit date: | 2007-03-07 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and dynamics of LC8 complexes with KXTQT-motif peptides: swallow and dynein intermediate chain compete for a common site.
J.Mol.Biol., 371, 2007
|
|
8F2H
 
 | |
8F2I
 
 | | P53 monomer structure | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Solares, M, Kelly, D.F. | | Deposit date: | 2022-11-08 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | High-Resolution Imaging of Human Cancer Proteins Using Microprocessor Materials.
Chembiochem, 23, 2022
|
|
6XRE
 
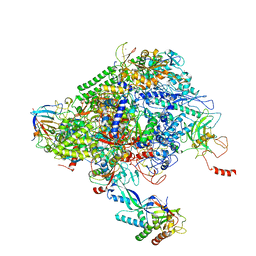 | | Structure of the p53/RNA polymerase II assembly | | Descriptor: | Cellular tumor antigen p53, DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11-a, ... | | Authors: | Liou, S.-H, Singh, S, Singer, R.H, Coleman, R.A, Liu, W. | | Deposit date: | 2020-07-12 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the p53/RNA polymerase II assembly.
Commun Biol, 4, 2021
|
|
6LA8
 
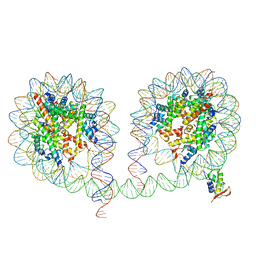 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Lee, P.L, Sharma, D, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6LA9
 
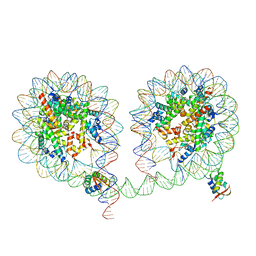 | | 349 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 (high cryoprotectant) | | Descriptor: | CALCIUM ION, DNA (349-MER), Histone H1.0, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Near-atomic resolution structures of interdigitated nucleosome fibres.
Nat Commun, 11, 2020
|
|
6LDM
 
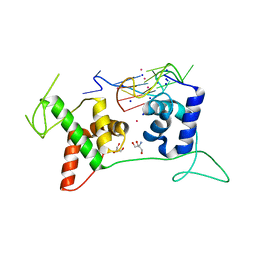 | | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA-binding protein RAP1, G-guadruplex DNA, ... | | Authors: | Traczyk, A, Gill, D.J, Chong, W.L, Rhodes, D. | | Deposit date: | 2019-11-22 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of G-quadruplex DNA recognition by the yeast telomeric protein Rap1.
Nucleic Acids Res., 48, 2020
|
|
6LAB
 
 | | 169 bp nucleosome, harboring cohesive DNA termini, assembled with linker histone H1.0 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (169-MER), ... | | Authors: | Adhireksan, Z, Sharma, D, Bao, Q, Lee, P.L, Padavattan, S, Davey, C.A. | | Deposit date: | 2019-11-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
6LA2
 
 | | 343 bp di-nucleosome harboring cohesive DNA termini assembled with linker histone H1.0 | | Descriptor: | DNA (343-MER), Histone H1.0, Histone H2A type 1-B/E, ... | | Authors: | Adhireksan, Z, Sharma, D, Lee, P.L, Davey, C.A. | | Deposit date: | 2019-11-11 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.89 Å) | | Cite: | Engineering nucleosomes for generating diverse chromatin assemblies.
Nucleic Acids Res., 49, 2021
|
|
