8WQN
 
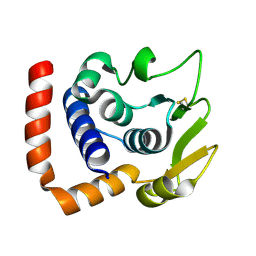 | |
8WQ8
 
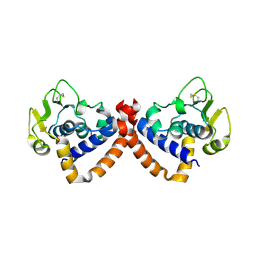 | |
7TB5
 
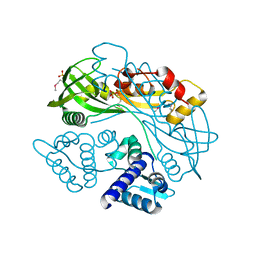 | | Structure of P. aeruginosa PA17 CapW | | Descriptor: | SULFATE ION, WYL domain-containing protein | | Authors: | Blankenchip, C.L, Nguyen, J.V, Lau, R.K, Ye, Q, Corbett, K.D. | | Deposit date: | 2021-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2022-06-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Control of bacterial immune signaling by a WYL domain transcription factor.
Nucleic Acids Res., 50, 2022
|
|
6T4B
 
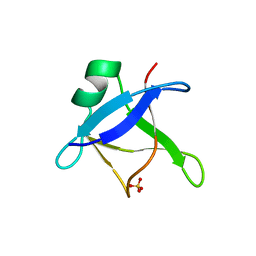 | | CRYSTAL STRUCTURE OF HUMAN TDP-43 N-TERMINAL DOMAIN AT 2.55 A RESOLUTION | | Descriptor: | SULFATE ION, TAR DNA-binding protein 43 | | Authors: | Watanabe, T.F, Wright, G.S.A, Amporndanai, K, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-10-13 | | Release date: | 2020-05-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Purification and Structural Characterization of Aggregation-Prone Human TDP-43 Involved in Neurodegenerative Diseases.
Iscience, 23, 2020
|
|
4A0C
 
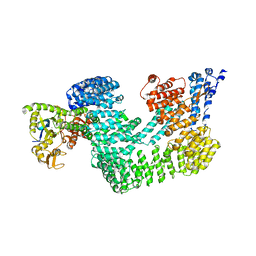 | | Structure of the CAND1-CUL4B-RBX1 complex | | Descriptor: | CULLIN-4B, CULLIN-ASSOCIATED NEDD8-DISSOCIATED PROTEIN 1, E3 UBIQUITIN-PROTEIN LIGASE RBX1, ... | | Authors: | Scrima, A, Fischer, E.S, Faty, M, Gut, H, Thoma, N.H. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The Molecular Basis of Crl4(Ddb2/Csa) Ubiquitin Ligase Architecture, Targeting, and Activation
Cell(Cambridge,Mass.), 147, 2011
|
|
8FD2
 
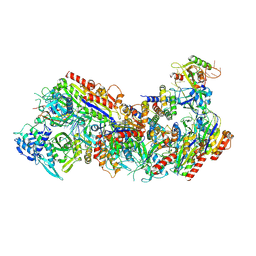 | | Cryo-EM structure of Cascade complex in type I-B CAST system | | Descriptor: | A Type I-B CRISPR-associated protein Cas5, RNA, Type I-B CRISPR-associated protein Cas11, ... | | Authors: | Chang, L, Wang, S. | | Deposit date: | 2022-12-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector.
Cell, 186, 2023
|
|
7SII
 
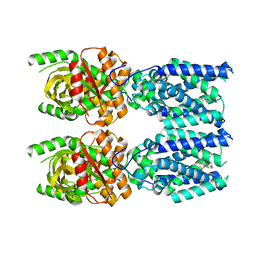 | | Human STING bound to both cGAMP and 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide (Compound 53) | | Descriptor: | 1-[(2-chloro-6-fluorophenyl)methyl]-3,3-dimethyl-2-oxo-N-[(2,4,6-trifluorophenyl)methyl]-2,3-dihydro-1H-indole-6-carboxamide, Stimulator of interferon genes protein, cGAMP | | Authors: | Lu, D, Shang, G, Jie, L, Lu, Y, Bai, X.C, Zhang, X. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Activation of STING by targeting a pocket in the transmembrane domain.
Nature, 604, 2022
|
|
3QP5
 
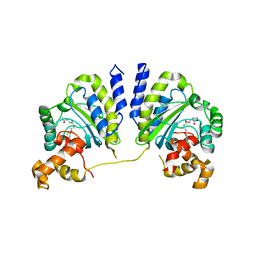 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
3O75
 
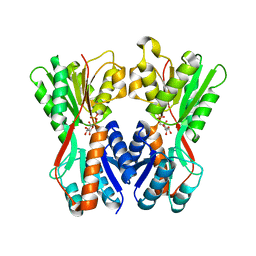 | | Crystal structure of Cra transcriptional dual regulator from Pseudomonas putida in complex with fructose 1-phosphate' | | Descriptor: | 1-O-phosphono-beta-D-fructofuranose, Fructose transport system repressor FruR | | Authors: | Chavarria, M, Santiago, C, Platero, R, Krell, T, Casasnovas, J.M, de Lorenzo, V. | | Deposit date: | 2010-07-30 | | Release date: | 2011-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fructose 1-phosphate is the preferred effector of the metabolic regulator Cra of Pseudomonas putida
J.Biol.Chem., 286, 2011
|
|
3E5Q
 
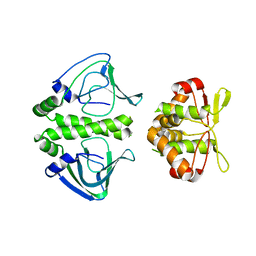 | | Unbound Oxidised CprK | | Descriptor: | Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
8FCX
 
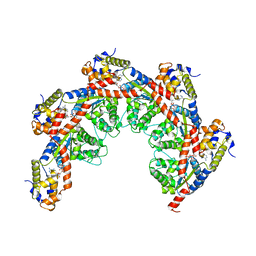 | |
8YK9
 
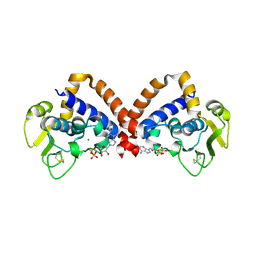 | | Structure of Saccharolobus solfataricus SegC (SSO0033) protein, ATP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SegC | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea.
Nucleic Acids Res., 52, 2024
|
|
1MNT
 
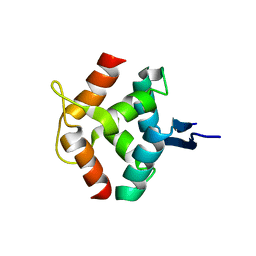 | | SOLUTION STRUCTURE OF DIMERIC MNT REPRESSOR (1-76) | | Descriptor: | MNT REPRESSOR | | Authors: | Burgering, M.J.M, Boelens, R, Gilbert, D.E, Breg, J.N, Knight, K.L, Sauer, R.T, Kaptein, R. | | Deposit date: | 1994-06-28 | | Release date: | 1994-09-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of dimeric Mnt repressor (1-76).
Biochemistry, 33, 1994
|
|
6TRI
 
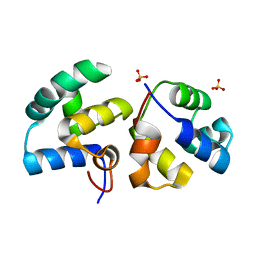 | | CI-MOR repressor-antirepressor complex of the temperate bacteriophage TP901-1 from Lactococcus lactis | | Descriptor: | CI, MOR, SULFATE ION | | Authors: | Rasmussen, K.K, Blackledge, M, Herrmann, T, Jensen, M.R, Lo Leggio, L. | | Deposit date: | 2019-12-18 | | Release date: | 2020-08-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.277 Å) | | Cite: | Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
2P14
 
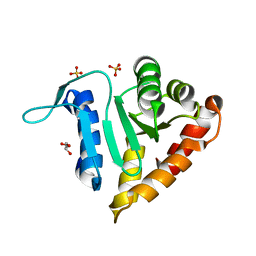 | | Crystal structure of small subunit (R.BspD6I2) of the heterodimeric restriction endonuclease R.BspD6I | | Descriptor: | GLYCEROL, Heterodimeric restriction endonuclease R.BspD6I small subunit, SULFATE ION | | Authors: | Kachalova, G.S, Bartunik, H.D, Artyukh, R.I, Rogulin, E.A, Yunusova, A.K, Zheleznaya, L.A, Matvienko, N.I. | | Deposit date: | 2007-03-02 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit.
J.Mol.Biol., 384, 2008
|
|
6RPR
 
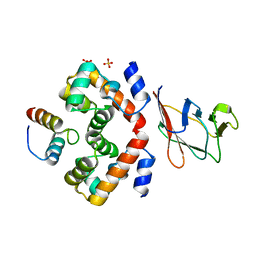 | |
7S4A
 
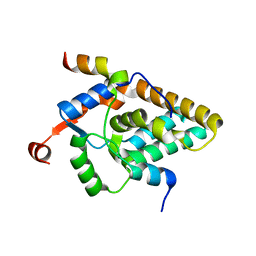 | | MRG15 complex with PALB2 peptide | | Descriptor: | Mortality factor 4-like protein 1, Partner and localizer of BRCA2 | | Authors: | Korolev, S, Deveryshetty, J. | | Deposit date: | 2021-09-08 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Structural Insight into the Mechanism of PALB2 Interaction with MRG15.
Genes (Basel), 12, 2021
|
|
3JTH
 
 | |
3QP6
 
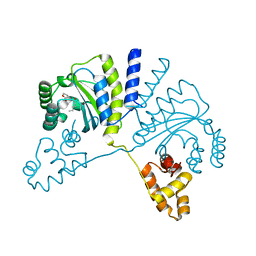 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
1OVF
 
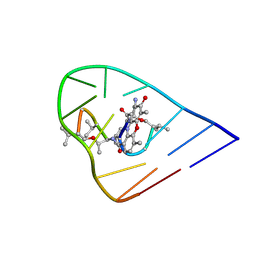 | | NMR Structure of ActD/5'-CCGTTTTGTGG-3' Complex | | Descriptor: | (5'-D(*CP*CP*GP*TP*TP*TP*TP*GP*TP*GP*G)-3'), ACTINOMYCIN D | | Authors: | Chin, K.-H, Chou, S.-H, Chen, F.-M. | | Deposit date: | 2003-03-26 | | Release date: | 2003-05-27 | | Last modified: | 2024-07-10 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Actd-5'-Ccgtt(3)Gtgg-3' Complex: Drug Interaction with Tandem G.T Mismatches and Hairpin Loop Backbone.
Nucleic Acids Res., 31, 2003
|
|
2BNM
 
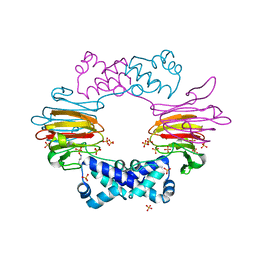 | |
1EA6
 
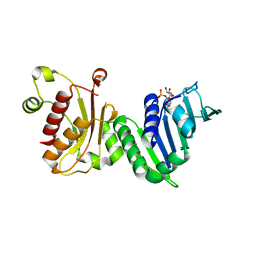 | |
7NFU
 
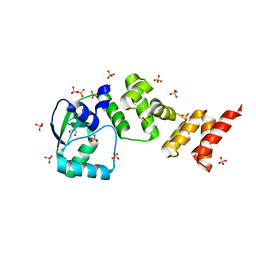 | | Crystal structure of C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | GLYCEROL, Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-17 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
7NG0
 
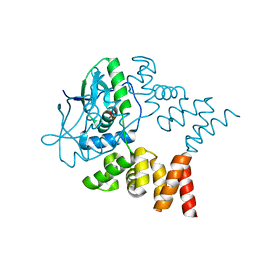 | | Crystal structure of N- and C-terminally truncated Geobacillus thermoleovorans nucleoid occlusion protein Noc | | Descriptor: | Nucleoid occlusion protein, SULFATE ION | | Authors: | Jalal, A.S.B, Tran, N.T, Wu, L.J, Ramakrishnan, K, Rejzek, M, Stevenson, C.E.M, Lawson, D.M, Errington, J, Le, T.B.K. | | Deposit date: | 2021-02-08 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | CTP regulates membrane-binding activity of the nucleoid occlusion protein Noc.
Mol.Cell, 81, 2021
|
|
5CJ9
 
 | | Bacillus halodurans Arginine repressor, ArgR | | Descriptor: | Arginine repressor, S-1,2-PROPANEDIOL | | Authors: | Park, Y.W, Kang, J, Yeo, H.Y, Lee, J.Y. | | Deposit date: | 2015-07-14 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.409 Å) | | Cite: | Structural Analysis and Insights into the Oligomeric State of an Arginine-Dependent Transcriptional Regulator from Bacillus halodurans.
Plos One, 11, 2016
|
|
