1XFK
 
 | |
6FFK
 
 | | Human apo-SOD1 bound to PtCl2(1R,2R-1,4-DACH | | Descriptor: | PtCl2(1(R),2(R)-DACH), Superoxide dismutase [Cu-Zn] | | Authors: | Calderone, V, Nativi, C, Cantini, F, Di Cesare Mannelli, L. | | Deposit date: | 2018-01-08 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Interaction of Half Oxa-/Halfcis-Platin Complex with Human Superoxide Dismutase and Induced Reduction of Neurotoxicity.
ACS Med Chem Lett, 9, 2018
|
|
4YSW
 
 | | Structure of rat xanthine oxidoreductase, C-terminal deletion protein variant, NADH bound form | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Nishino, T, Okamoto, K, Kawaguchi, Y, Matsumura, T, Eger, B.T, Pai, E.F. | | Deposit date: | 2015-03-17 | | Release date: | 2015-04-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The C-terminal peptide plays a role in the formation of an intermediate form during the transition between xanthine dehydrogenase and xanthine oxidase.
Febs J., 282, 2015
|
|
5JMM
 
 | |
7DNF
 
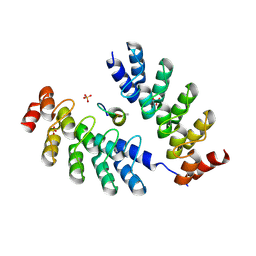 | | DARPin 63_B7 in complex with V3-IY (MN) crown mimetic | | Descriptor: | DARPin 63_B7, SULFATE ION, V3-IY (MN) crown mimetic peptide | | Authors: | Wu, Y, Plueckthun, A. | | Deposit date: | 2020-12-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Nat Commun, 12, 2021
|
|
5EPR
 
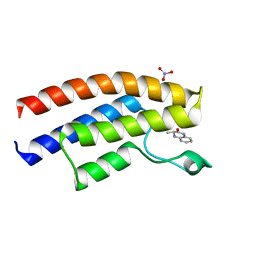 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED11 | | Descriptor: | (3~{S})-3-methyl-3,4-dihydro-1~{H}-quinoxalin-2-one, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
4NFY
 
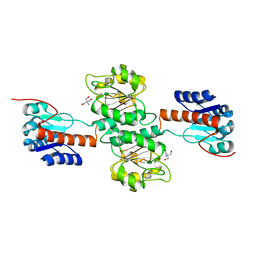 | |
5EI1
 
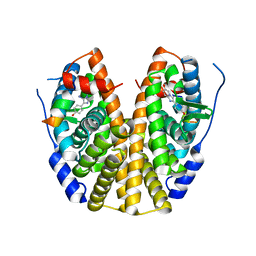 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the imidazopyridine derivative 2-(4-hydroxyphenyl)-3-iodanyl-imidazo[1,2-a]pyridin-6-ol | | Descriptor: | 2-(4-hydroxyphenyl)-3-iodanyl-imidazo[1,2-a]pyridin-6-ol, Estrogen receptor, NCOA2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-10-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5HEX
 
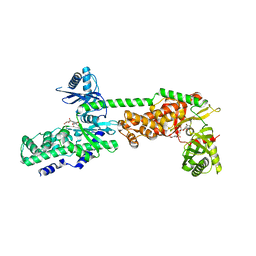 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5I9Y
 
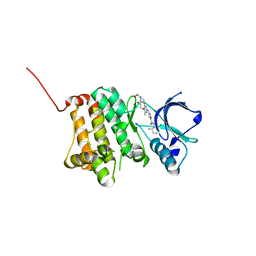 | | Crystal Structure of Ephrin A2 (EphA2) Receptor Protein Kinase with dasatinib | | Descriptor: | 1,2-ETHANEDIOL, Ephrin type-A receptor 2, N-(2-CHLORO-6-METHYLPHENYL)-2-({6-[4-(2-HYDROXYETHYL)PIPERAZIN-1-YL]-2-METHYLPYRIMIDIN-4-YL}AMINO)-1,3-THIAZOLE-5-CARBOXAMIDE | | Authors: | Kudlinzki, D, Linhard, V.L, Gande, S.L, Sreeramulu, S, Saxena, K, Heinzlmeir, S, Medard, G, Kuester, B, Schwalbe, H. | | Deposit date: | 2016-02-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.228 Å) | | Cite: | Chemical Proteomics and Structural Biology Define EPHA2 Inhibition by Clinical Kinase Drugs.
ACS Chem. Biol., 11, 2016
|
|
7K73
 
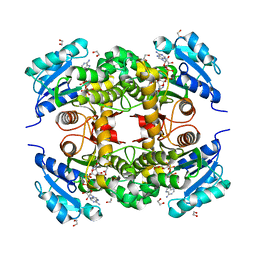 | |
5KDR
 
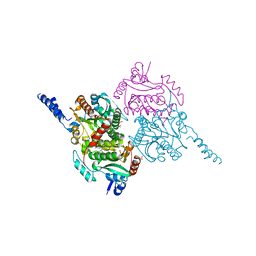 | | The crystal structure of carboxyltransferase from Staphylococcus Aureus bound to the antimicrobial agent moiramide B. | | Descriptor: | 1,2-ETHANEDIOL, Acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, Acetyl-coenzyme A carboxylase carboxyl transferase subunit beta, ... | | Authors: | Silvers, M.A, Pakhomova, S, Neau, D, Silvers, W.C, Anzalone, N, Taylor, C.M, Waldrop, G.L. | | Deposit date: | 2016-06-08 | | Release date: | 2016-08-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Carboxyltransferase from Staphylococcus aureus Bound to the Antibacterial Agent Moiramide B.
Biochemistry, 55, 2016
|
|
6TCL
 
 | | Photosystem I tetramer | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Chen, M, Perez-Boerema, A, Li, S, Amunts, A. | | Deposit date: | 2019-11-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Distinct structural modulation of photosystem I and lipid environment stabilizes its tetrameric assembly.
Nat.Plants, 6, 2020
|
|
8J6Z
 
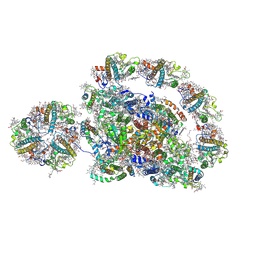 | | Cryo-EM structure of the Arabidopsis thaliana photosystem I(PSI-LHCII-ST2) | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chen, S.J.B, Wu, J.H, Sui, S.F, Zhang, L.X. | | Deposit date: | 2023-04-26 | | Release date: | 2023-11-15 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Regulatory dynamics of the higher-plant PSI-LHCI supercomplex during state transitions.
Mol Plant, 16, 2023
|
|
6BJP
 
 | |
9FSN
 
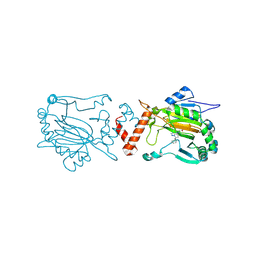 | | FIH in complex with Enarodustat crystal structure at 2.2A | | Descriptor: | Enarodustat, Hypoxia-inducible factor 1-alpha inhibitor, MANGANESE (II) ION | | Authors: | Corner, T, Brewitz, L, Kaur, S, Schnaubelt, L.I, Allen, M.D, Schofield, C.J. | | Deposit date: | 2024-06-21 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic and Selectivity Studies on the Approved HIF Prolyl Hydroxylase Inhibitors Desidustat and Enarodustat.
Chemmedchem, 19, 2024
|
|
6KDY
 
 | |
5ELO
 
 | |
8IHZ
 
 | | FACTOR INHIBITING HIF-1 ALPHA in complex with (5-(1-(3-(4-chlorophenyl)propyl)-1H-1,2,3-triazol-4-yl)-3-hydroxypicolinoyl)glycine | | Descriptor: | 2-[[5-[1-[3-(4-chlorophenyl)propyl]-1,2,3-triazol-4-yl]-3-oxidanyl-pyridin-2-yl]carbonylamino]ethanoic acid, Hypoxia-inducible factor 1-alpha inhibitor, SULFATE ION, ... | | Authors: | Nakashima, Y, Corner, T, Zhang, X, Schofield, C.J. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | A Small-Molecule Inhibitor of Factor Inhibiting HIF Binding to a Tyrosine-Flip Pocket for the Treatment of Obesity.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
4NOH
 
 | | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis. | | Descriptor: | CHLORIDE ION, Lipoprotein, putative | | Authors: | Minasov, G, Shuvalova, L, Halavaty, A, Winsor, J, Dubrovska, I, Shatsman, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.502 Å) | | Cite: | 1.5 Angstrom Crystal Structure of Putative Lipoprotein from Bacillus anthracis.
TO BE PUBLISHED
|
|
4BFL
 
 | |
5EQ1
 
 | | Crystal structure of the human BRPF1 bromodomain in complex with SEED12 | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NITRATE ION, Peregrin | | Authors: | Zhu, J, Caflisch, A. | | Deposit date: | 2015-11-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Twenty Crystal Structures of Bromodomain and PHD Finger Containing Protein 1 (BRPF1)/Ligand Complexes Reveal Conserved Binding Motifs and Rare Interactions.
J.Med.Chem., 59, 2016
|
|
6BZ5
 
 | |
4ARV
 
 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
5UX6
 
 | | Structure of Human POFUT1 in its apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL | | Authors: | Xu, X, McMillan, B, Blacklow, S.C. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human POFUT1, its requirement in ligand-independent oncogenic Notch signaling, and functional effects of Dowling-Degos mutations.
Glycobiology, 27, 2017
|
|
