8BBS
 
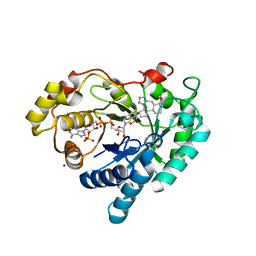 | | Structure of AKR1C3 in complex with a bile acid fused tetrazole inhibitor | | Descriptor: | (4~{R})-4-[(1~{R},2~{S},5~{R},6~{R},13~{S},14~{S},17~{R},19~{R})-6,14-dimethyl-17-oxidanyl-7,8,9,10-tetrazapentacyclo[11.8.0.0^{2,6}.0^{7,11}.0^{14,19}]henicosa-8,10-dien-5-yl]pentanoic acid, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Petri, E.T, Skerlova, J, Marinovic, M, Brynda, J, Kugler, M, Skoric, D, Bekic, S, Celic, A.S, Rezacova, P. | | Deposit date: | 2022-10-14 | | Release date: | 2023-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray structure of human aldo-keto reductase 1C3 in complex with a bile acid fused tetrazole inhibitor: experimental validation, molecular docking and structural analysis.
Rsc Med Chem, 14, 2023
|
|
6CZ9
 
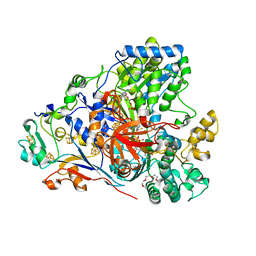 | | The arsenate respiratory reductase (Arr) complex from Shewanella sp. ANA-3 bound to arsenite | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, 4Fe-4S ferredoxin, ... | | Authors: | Glasser, N.R, Newman, D.K. | | Deposit date: | 2018-04-08 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic analysis of the arsenate respiratory reductase provides insight into environmental arsenic transformations.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5XPN
 
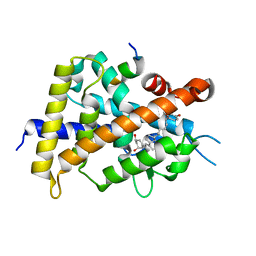 | | Crystal structure of VDR-LBD complexed with 25RS-(hydroxyphenyl)-25-methoxy-2-methylidene-19,26,27-trinor-1-hydroxyvitamin D3 | | Descriptor: | (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{R})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, (1~{R},3~{R})-5-[(2~{E})-2-[(1~{R},3~{a}~{S},7~{a}~{R})-1-[(2~{R},6~{S})-6-(4-hydroxyphenyl)-6-methoxy-hexan-2-yl]-7~{a}-methyl-2,3,3~{a},5,6,7-hexahydro-1~{H}-inden-4-ylidene]ethylidene]-2-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, ... | | Authors: | Kato, A, Itoh, T, Yamamoto, K. | | Deposit date: | 2017-06-03 | | Release date: | 2018-07-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Vitamin D Analogues with a p-Hydroxyphenyl Group at the C25 Position: Crystal Structure of Vitamin D Receptor Ligand-Binding Domain Complexed with the Ligand Explains the Mechanism Underlying Full Antagonistic Action
J. Med. Chem., 60, 2017
|
|
5TPI
 
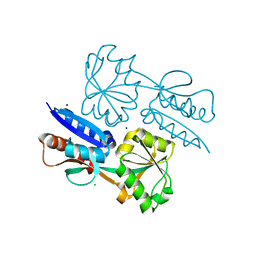 | | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator (LysR family), SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.
To Be Published
|
|
6HRO
 
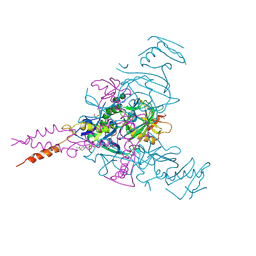 | | Crystal structure of Ebolavirus glycoprotein in complex with inhibitor 118a | | Descriptor: | 1-[2-[4-[4-(4-chlorophenyl)-3-methyl-1~{H}-pyrazol-5-yl]-3-oxidanyl-phenoxy]ethyl]piperidin-1-ium-4-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Ren, J, Zhao, Y, Stuart, D.I. | | Deposit date: | 2018-09-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based in Silico Screening Identifies a Potent Ebolavirus Inhibitor from a Traditional Chinese Medicine Library.
J.Med.Chem., 62, 2019
|
|
5UJU
 
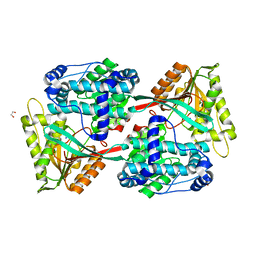 | |
4L3H
 
 | |
5XDX
 
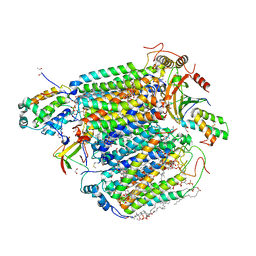 | | Bovine heart cytochrome c oxidase in the reduced state with pH 7.3 at 1.99 angstrom resolution | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Luo, F.J, Shimada, A, Hagimoto, N, Shimada, S, Shinzawa-Itoh, K, Yamashita, E, Yoshikawa, S, Tsukihara, T. | | Deposit date: | 2017-03-30 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of bovine cytochrome c oxidase in the ligand-free reduced state at neutral pH.
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
5DXQ
 
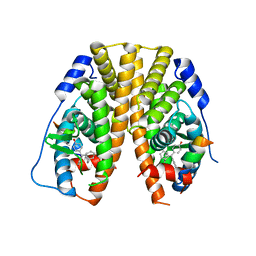 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with the Cyclofenil Derivative 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol | | Descriptor: | 4,4'-[(1s,5s)-bicyclo[3.3.1]non-9-ylidenemethanediyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-23 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3PDX
 
 | | Crystal structural of mouse tyrosine aminotransferase | | Descriptor: | Tyrosine aminotransferase | | Authors: | Mehere, P.V, Han, Q, Lemkul, J.A, Robinson, H, Bevan, D.R, Li, J. | | Deposit date: | 2010-10-25 | | Release date: | 2010-11-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Tyrosine aminotransferase: biochemical and structural properties and molecular dynamics simulations.
Protein Cell, 1, 2010
|
|
6HU9
 
 | | III2-IV2 mitochondrial respiratory supercomplex from S. cerevisiae | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, 5-(3,7,11,15,19,23-HEXAMETHYL-TETRACOSA-2,6,10,14,18,22-HEXAENYL)-2,3-DIMETHOXY-6-METHYL-BENZENE-1,4-DIOL, CALCIUM ION, ... | | Authors: | Hartley, A.M, Pinotsis, N, Marechal, A. | | Deposit date: | 2018-10-05 | | Release date: | 2018-12-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat. Struct. Mol. Biol., 26, 2019
|
|
5SX5
 
 | |
5T0L
 
 | | Crystal structure of Toxoplasma gondii TS-DHFR complexed with NADPH, dUMP, PDDF and 5-(4-(3,4-dichlorophenyl)piperazin-1-yl)pyrimidine-2,4-diamine (TRC-15) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 5-[4-(3,4-dichlorophenyl)piperazin-1-yl]pyrimidine-2,4-diamine, ... | | Authors: | Thomas, S.B, Chen, Z, Lu, H, Li, Y. | | Deposit date: | 2016-08-16 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Discovery of Potent and Selective Leads against Toxoplasma gondii Dihydrofolate Reductase via Structure-Based Design.
ACS Med Chem Lett, 7, 2016
|
|
4YSZ
 
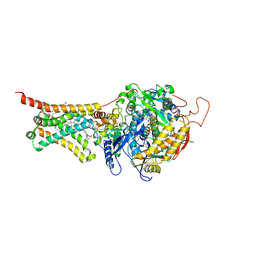 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide | | Descriptor: | 2-iodo-N-[3-(1-methylethoxy)phenyl]benzamide, Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Int J Mol Sci, 16, 2015
|
|
6DDP
 
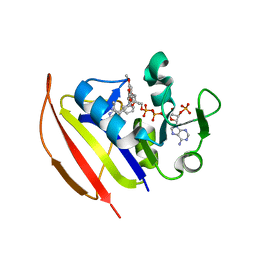 | | Mycobacterium tuberculosis Dihydrofolate Reductase complexed with beta-NADPH and 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid | | Descriptor: | 3'-[(2R)-4-(2,4-diamino-6-ethylpyrimidin-5-yl)but-3-yn-2-yl]-5'-methoxy[1,1'-biphenyl]-4-carboxylic acid, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hajian, B, Wright, D. | | Deposit date: | 2018-05-10 | | Release date: | 2018-05-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Drugging the Folate Pathway in Mycobacterium tuberculosis: The Role of Multi-targeting Agents.
Cell Chem Biol, 26, 2019
|
|
8AWR
 
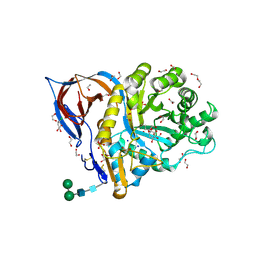 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl chloride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-chloranylcyclohex-4-ene-1,2,3-triol, (1~{S},2~{S},3~{S},4~{R})-cyclohexane-1,2,3,4-tetrol, 1,2-ETHANEDIOL, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
8AX3
 
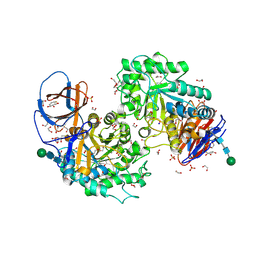 | | Structure of recombinant human beta-glucocerebrosidase in complex with L-carbaxylosyl fluoride | | Descriptor: | (1~{S},2~{R},3~{S},6~{S})-6-fluoranylcyclohex-4-ene-1,2,3-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Davies, G.J. | | Deposit date: | 2022-08-30 | | Release date: | 2024-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Single turnover covalent inhibitors for functional chaperoning of lysosomal glycoside hydrolases
To be published
|
|
4YWJ
 
 | |
1LUD
 
 | | SOLUTION STRUCTURE OF DIHYDROFOLATE REDUCTASE COMPLEXED WITH TRIMETHOPRIM AND NADPH, 24 STRUCTURES | | Descriptor: | 2,4-DIAMINO-5-(3,4,5-TRIMETHOXY-BENZYL)-PYRIMIDIN-1-IUM, DIHYDROFOLATE REDUCTASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Polshakov, V.I, Smirnov, E.G, Birdsall, B, Kelly, G, Feeney, J. | | Deposit date: | 2002-05-22 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR-based solution structure of the complex of Lactobacillus casei dihydrofolate reductase with trimethoprim and NADPH
J.Biomol.Nmr, 24, 2002
|
|
6E62
 
 | | Crystal structure of malaria transmission-blocking antigen Pfs48/45 6C in complex with antibody 85RF45.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 85RF45.1 Fab heavy chain, 85RF45.1 Fab light chain, ... | | Authors: | Kundu, P, Semesi, A, Julien, J.P. | | Deposit date: | 2018-07-23 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural delineation of potent transmission-blocking epitope I on malaria antigen Pfs48/45.
Nat Commun, 9, 2018
|
|
5EJW
 
 | | Crystal structure of chromobox homolog 7 (CBX7) chromodomain with MS351 | | Descriptor: | (1~{R})-2-[2-azanylidene-3-[(2-methylphenyl)methyl]benzimidazol-1-yl]-1-(3,4-dichlorophenyl)ethanol, Chromobox protein homolog 7 | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2015-11-02 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Guided Discovery of Selective Antagonists for the Chromodomain of Polycomb Repressive Protein CBX7.
Acs Med.Chem.Lett., 7, 2016
|
|
6MBJ
 
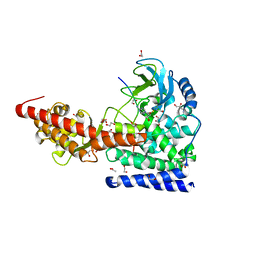 | | SETD3, a Histidine Methyltransferase, in Complex with an Actin Peptide and SAH, P21 Crystal Form | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Actin Peptide, ... | | Authors: | Horton, J.R, Dai, S, Cheng, X. | | Deposit date: | 2018-08-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | SETD3 is an actin histidine methyltransferase that prevents primary dystocia.
Nature, 565, 2019
|
|
5K2C
 
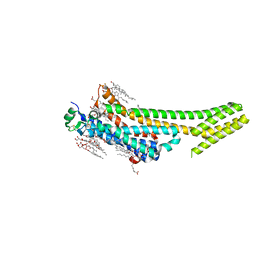 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
3ML1
 
 | | Crystal Structure of the Periplasmic Nitrate Reductase from Cupriavidus necator | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIOXOTHIOMOLYBDENUM(VI) ION, Diheme cytochrome c napB, ... | | Authors: | Coelho, C, Trincao, J, Romao, M.J. | | Deposit date: | 2010-04-16 | | Release date: | 2011-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of Cupriavidus necator nitrate reductase in oxidized and partially reduced states
J.Mol.Biol., 408, 2011
|
|
1SFS
 
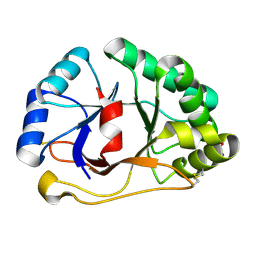 | | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein | | Descriptor: | Hypothetical protein, PHOSPHATE ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Moy, S.F, Collart, F.R, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | 1.07 A crystal structure of an uncharacterized B. stearothermophilus protein
To be Published
|
|
