2G38
 
 | | A PE/PPE Protein Complex from Mycobacterium tuberculosis | | Descriptor: | MANGANESE (II) ION, PE FAMILY PROTEIN, PPE FAMILY PROTEIN | | Authors: | Strong, M, Sawaya, M.R, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2GDN
 
 | |
2G7N
 
 | | Structure of the Light Chain of Botulinum Neurotoxin Serotype A Bound to small Molecule Inhibitors | | Descriptor: | Botulinum neurotoxin type A, SILVER ION, ZINC ION | | Authors: | Fu, Z, Baldwin, M.R, Boldt, G.E, Janda, K.D, Barbieri, J.T, Kim, J.-J.P. | | Deposit date: | 2006-02-28 | | Release date: | 2007-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the Light Chain of Botulinum Neurotoxin Serotype A bound to Small Molecule Inhibitors
To be Published
|
|
2G44
 
 | | Human Estrogen Receptor Alpha Ligand-Binding Domain In Complex With OBCP-1M-G and A Glucocorticoid Receptor Interacting Protein 1 NR Box II Peptide | | Descriptor: | 4-[(1S,2R,5S)-4,4,8-TRIMETHYL-3-OXABICYCLO[3.3.1]NON-7-EN-2-YL]PHENOL, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Rajan, S.S, Hsieh, R.W, Sharma, S.K, Greene, G.L. | | Deposit date: | 2006-02-21 | | Release date: | 2007-01-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and characterization of novel estrogen
receptor agonist ligands and development of biochips for
nuclear receptor drug discovery
Thesis, 2006
|
|
2G8N
 
 | | Structure of hPNMT with inhibitor 3-Hydroxymethyl-7-(N-4-chlorophenylaminosulfonyl)-THIQ and AdoHcy | | Descriptor: | (3R)-N-(4-CHLOROPHENYL)-3-(HYDROXYMETHYL)-1,2,3,4-TETRAHYDROISOQUINOLINE-7-SULFONAMIDE, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Gee, C.L, Martin, J.L. | | Deposit date: | 2006-03-02 | | Release date: | 2006-09-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Comparison of the Binding of 3-Fluoromethyl-7-sulfonyl-1,2,3,4-tetrahydroisoquinolines with Their Isosteric Sulfonamides to the Active Site of Phenylethanolamine N-Methyltransferase
J.Med.Chem., 49, 2006
|
|
2JPA
 
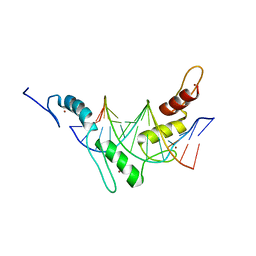 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | Descriptor: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | Authors: | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2007-05-01 | | Release date: | 2007-10-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
2JR7
 
 | | Solution structure of human DESR1 | | Descriptor: | DPH3 homolog, ZINC ION | | Authors: | Wu, F, Wu, J, Shi, Y. | | Deposit date: | 2007-06-21 | | Release date: | 2008-03-25 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human DESR1, a CSL zinc-binding protein
Proteins, 71, 2008
|
|
2JE0
 
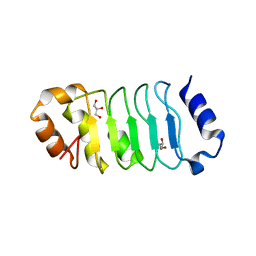 | | Crystal Structure of pp32 | | Descriptor: | ACIDIC LEUCINE-RICH NUCLEAR PHOSPHOPROTEIN 32 FAMILY MEMBER A, GLYCEROL | | Authors: | Huyton, T, Wolberger, C. | | Deposit date: | 2007-01-12 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of the Tumor Suppressor Protein Pp32 (Anp32A): Structural Insights Into Anp32 Family of Proteins.
Protein Sci., 16, 2007
|
|
2JE1
 
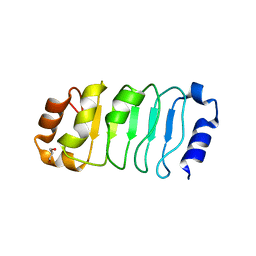 | |
2JJM
 
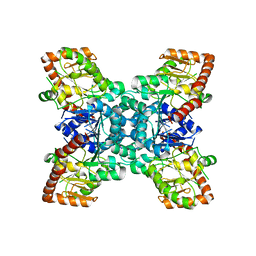 | |
2LO4
 
 | |
2L16
 
 | |
2LB0
 
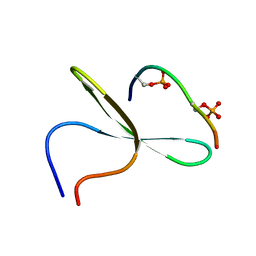 | | Structure of the first WW domain of human Smurf1 in complex with a di-phosphorylated human Smad1 derived peptide | | Descriptor: | E3 ubiquitin-protein ligase SMURF1, Mothers against decapentaplegic homolog 1 | | Authors: | Macias, M.J, Aragon, E, Goerner, N, Zaromytidou, A, Xi, Q, Escobedo, A, Massague, J. | | Deposit date: | 2011-03-22 | | Release date: | 2011-07-06 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | A Smad action turnover switch operated by WW domain readers of a phosphoserine code.
Genes Dev., 25, 2011
|
|
2L4Z
 
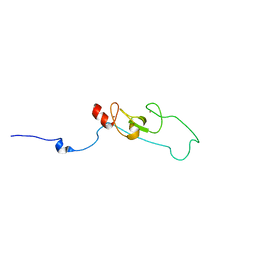 | | NMR structure of fusion of CtIP (641-685) to LMO4-LIM1 (18-82) | | Descriptor: | DNA endonuclease RBBP8,LIM domain transcription factor LMO4, ZINC ION | | Authors: | Liew, C, Stokes, P.H, Kwan, A.H, Matthews, J.M. | | Deposit date: | 2010-10-22 | | Release date: | 2011-10-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Interaction of the Breast Cancer Oncogene LMO4 with the Tumour Suppressor CtIP/RBBP8.
J.Mol.Biol., 425, 2013
|
|
2L28
 
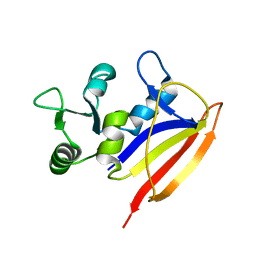 | | Solution structure of lactobacillus casei dihydrofolate reductase apo-form, 25 conformers | | Descriptor: | Dihydrofolate reductase | | Authors: | Polshakov, V.I, Birdsall, B, Feeney, J. | | Deposit date: | 2010-08-13 | | Release date: | 2011-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Apo L. casei Dihydrofolate Reductase and Its Complexes with Trimethoprim and NADPH: Contributions to Positive Cooperative Binding from Ligand-Induced Refolding, Conformational Changes, and Interligand Hydrophobic Interactions.
Biochemistry, 50, 2011
|
|
2LA4
 
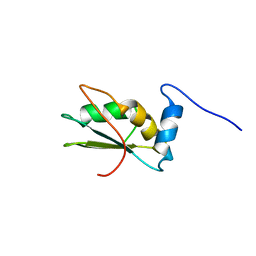 | | NMR structure of the C-terminal RRM domain of poly(U) binding 1 | | Descriptor: | Nuclear and cytoplasmic polyadenylated RNA-binding protein PUB1 | | Authors: | Santiveri, C.M, Mirassou, Y, Rico-Lastres, P, Martinez-Lumbreras, S, Perez-Canadillas, J.M. | | Deposit date: | 2011-03-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Pub1p C-terminal RRM domain interacts with Tif4631p through a conserved region neighbouring the Pab1p binding site
Plos One, 6, 2011
|
|
2LCV
 
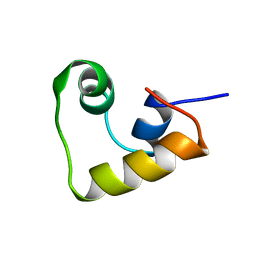 | |
2L7A
 
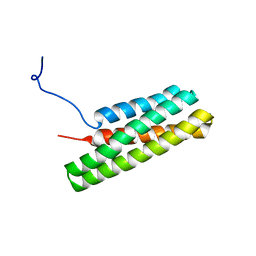 | | Solution Structure of the R3 Domain of Talin | | Descriptor: | Talin-1 | | Authors: | Goult, B.T, Gingras, A.R, Bate, N, Roberts, G.C.K, Barsukov, I.L, Critchley, D.R. | | Deposit date: | 2010-12-06 | | Release date: | 2011-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover.
J.Biol.Chem., 288, 2013
|
|
2LGJ
 
 | | Solution structure of MsPTH | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Yadav, R, Pathak, P, Pulavarti, S, Jain, A, Kumar, A, Shukla, V, Arora, A. | | Deposit date: | 2011-07-27 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of Peptidyl t-RNA hydrolase from Mycobacterium smegmatis
To be Published
|
|
2L1W
 
 | |
2LCK
 
 | |
2L18
 
 | |
2LEX
 
 | | Complex of the C-terminal WRKY domain of AtWRKY4 and a W-box DNA | | Descriptor: | DNA (5'-D(*CP*G*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*C*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*CP*G)-3'), Probable WRKY transcription factor 4, ... | | Authors: | Yamasaki, K, Kigawa, T, Watanabe, S, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-24 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence-spscific DNA recognition by an Arabidopsis WRKY transcription factor
J.Biol.Chem., 2012
|
|
2L19
 
 | | An arsenate reductase in the intermediate state | | Descriptor: | Arsenate reductase | | Authors: | Yu, C, Xia, B, Jin, C. | | Deposit date: | 2010-07-26 | | Release date: | 2011-04-13 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | (1)H, (13)C and (15)N resonance assignments of the arsenate reductase from Synechocystis sp. strain PCC 6803
Biomol.Nmr Assign., 5, 2011
|
|
2LNW
 
 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
