6EZ4
 
 | | NMR structure of the C-terminal domain of the human RPAP3 protein | | Descriptor: | RNA polymerase II-associated protein 3 | | Authors: | Fabre, P, Chagot, M.E, Bragantini, B, Manival, X, Quinternet, M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | The RPAP3-Cterminal domain identifies R2TP-like quaternary chaperones.
Nat Commun, 9, 2018
|
|
6PKJ
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.17A resolution (#2) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKO
 
 | | MicroED structure of proteinase K from a platinum coated, unpolished, single lamella at 2.07A resolution (#12) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.07 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKN
 
 | | MicroED structure of proteinase K from an unpolished, platinum-coated, single lamella at 2.08A resolution (#9) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.08 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKR
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.79A resolution (#13) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.79 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKK
 
 | | MicroED structure of proteinase K from an uncoated, single lamella at 2.18A resolution (#5) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.176 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
6PKP
 
 | | MicroED structure of proteinase K from a platinum-coated, polished, single lamella at 1.91A resolution (#10) | | Descriptor: | Proteinase K | | Authors: | Martynowycz, M.W, Zhao, W, Hattne, J, Jensen, G.J, Gonen, T. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-04 | | Last modified: | 2019-12-18 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.91 Å) | | Cite: | Qualitative Analyses of Polishing and Precoating FIB Milled Crystals for MicroED.
Structure, 27, 2019
|
|
7S7B
 
 | |
7S7C
 
 | |
9B73
 
 | | Cryo-EM structure of the desensitised ATP-bound human P2X1 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Felix, M.B, Alisa, G, Hariprasad, V, Jesse, I.M, David, M.T. | | Deposit date: | 2024-03-27 | | Release date: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (1.96 Å) | | Cite: | Structural insights into the human P2X1 receptor and ligand interactions
Nat Commun, 15, 2024
|
|
8QPY
 
 | |
8SWF
 
 | | Cryo-EM structure of NLRP3 open octamer | | Descriptor: | NACHT, LRR and PYD domains-containing protein 3 | | Authors: | Yu, X, Matico, R.E, Miller, R, Schoubroeck, B.V, Grauwen, K, Suarez, J, Pietrak, B, Haloi, N, Yin, Y, Tresadern, G.J, Perez-Benito, L, Lindahl, E, Bottelbergs, A, Oehlrich, D, Opdenbosch, N.V, Sharma, S. | | Deposit date: | 2023-05-18 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Cryo-EM structures of NLRP3 reveal its self-activation mechanism
Nat Commun, 2024
|
|
1ACC
 
 | | ANTHRAX PROTECTIVE ANTIGEN | | Descriptor: | ANTHRAX PROTECTIVE ANTIGEN, CALCIUM ION | | Authors: | Petosa, C, Liddington, R.C. | | Deposit date: | 1997-02-05 | | Release date: | 1998-02-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the anthrax toxin protective antigen.
Nature, 385, 1997
|
|
1D6G
 
 | |
6DU8
 
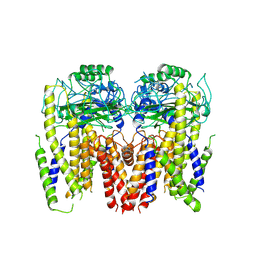 | | Human Polycsytin 2-l1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Polycystic kidney disease 2-like 1 protein | | Authors: | Hulse, R.E, Clapham, D.E, Li, Z, Huang, R.K, Zhang, J. | | Deposit date: | 2018-06-20 | | Release date: | 2018-07-25 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Cryo-EM structure of the polycystin 2-l1 ion channel.
Elife, 7, 2018
|
|
6D9O
 
 | |
7TYZ
 
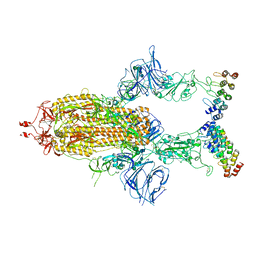 | | Cryo-EM structure of SARS-CoV-2 spike in complex with FSR22, an anti-SARS-CoV-2 DARPin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DARPin FSR22, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-02-15 | | Release date: | 2022-12-07 | | Last modified: | 2023-03-15 | | Method: | ELECTRON MICROSCOPY (3.51 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
6DLW
 
 | | Complement component polyC9 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement component C9, beta-D-mannopyranose | | Authors: | Dunstone, M.A, Spicer, B.A, Law, R.H.P. | | Deposit date: | 2018-06-03 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | The first transmembrane region of complement component-9 acts as a brake on its self-assembly.
Nat Commun, 9, 2018
|
|
6KI0
 
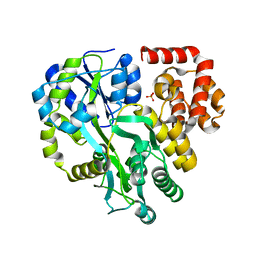 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
6DT6
 
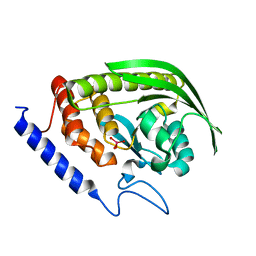 | | Crystal Structure of the YopH PTP1B Chimera 3 PTPase bound to vanadate | | Descriptor: | Targeted effector protein, VANADATE ION | | Authors: | Morales, Y, Johnson, S.J, Hengge, A.C. | | Deposit date: | 2018-06-15 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | A YopH PTP1B Chimera Shows the Importance of the WPD-Loop Sequence to the Activity, Structure, and Dynamics of Protein Tyrosine Phosphatases.
Biochemistry, 57, 2018
|
|
6DB5
 
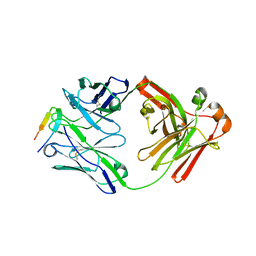 | |
6DBV
 
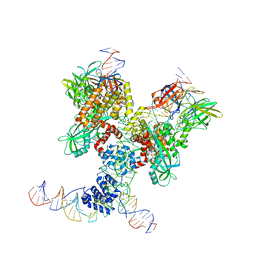 | | Cryo-EM structure of RAG in complex with 12-RSS and 23-RSS substrate DNAs | | Descriptor: | CALCIUM ION, Forward strand of 12-RSS substrate DNA, Forward strand of 23-RSS substrate DNA, ... | | Authors: | Wu, H, Liao, M, Ru, H, Mi, W. | | Deposit date: | 2018-05-03 | | Release date: | 2018-08-01 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.291 Å) | | Cite: | DNA melting initiates the RAG catalytic pathway.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6KNA
 
 | |
6K8Q
 
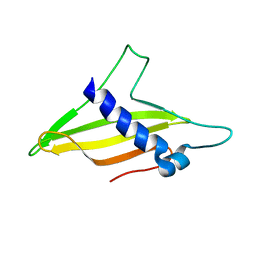 | | Solution structure of the intermembrane space domain of the mitochondrial import protein Tim21 from S. cerevisiae | | Descriptor: | Mitochondrial import inner membrane translocase subunit TIM21 | | Authors: | Bala, S, Shinya, S, Srivastava, A, Shimada, A, Kobayashi, N, Kojima, C, Tama, F, Miyashita, O, Kohda, D. | | Deposit date: | 2019-06-13 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Crystal contact-free conformation of an intrinsically flexible loop in protein crystal: Tim21 as the case study.
Biochim Biophys Acta Gen Subj, 1864, 2020
|
|
6K2K
 
 | | Solution structure of MUL1-RING domain | | Descriptor: | Mitochondrial ubiquitin ligase activator of NFKB 1, ZINC ION | | Authors: | Lee, M.S, Lee, M.K, Ryu, K.S, Chi, S.W. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of MUL1-RING domain and its interaction with p53 transactivation domain.
Biochem.Biophys.Res.Commun., 516, 2019
|
|
