6P4W
 
 | |
5MF0
 
 | | Crystal structure of Smad4-MH1 bound to the GGCCG site. | | Descriptor: | CHLORIDE ION, DNA (5'-D(P*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*GP*CP*CP*CP*GP*T)-3'), MH1 domain of human Smad4, ... | | Authors: | Kaczmarska, Z, Freier, R, Marquez, J.A, Macias, M.J. | | Deposit date: | 2016-11-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Structural basis for genome wide recognition of 5-bp GC motifs by SMAD transcription factors.
Nat Commun, 8, 2017
|
|
1GRJ
 
 | |
2W29
 
 | | Gly102Thr mutant of Rv3291c | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Dey, S, Ravishankar, R. | | Deposit date: | 2008-10-25 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
2P52
 
 | |
5YHX
 
 | | Structure of Lactococcus lactis ZitR, wild type | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
6P4O
 
 | |
6SJ7
 
 | |
5YI0
 
 | | Structure of Lactococcus lactis ZitR, C30AH42A mutant | | Descriptor: | ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
5GTC
 
 | | Crystal structure of complex between DMAP-SH conjugated with a Kaposi's sarcoma herpesvirus LANA peptide (5-15) and nucleosome core particle | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1-B/E, ... | | Authors: | Arimura, Y, Kato, D, Suto, H, Kurumizaka, H, Kawashima, S.A, Yamatsugu, K, Kanai, M. | | Deposit date: | 2016-08-19 | | Release date: | 2017-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Posttranslational Modifications: Chemical Catalyst-Driven Regioselective Histone Acylation of Native Chromatin.
J. Am. Chem. Soc., 139, 2017
|
|
4ZWT
 
 | |
5YI1
 
 | | Structure of Lactococcus lactis ZitR, C30AH42A mutant in apo form | | Descriptor: | Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
6Y8P
 
 | | Crystal structure of SNAP-tag labeled with a benzyl-tetramethylrhodamine fluorophore | | Descriptor: | 1,2-ETHANEDIOL, O6-alkylguanine-DNA alkyltransferase mutant, ZINC ION, ... | | Authors: | Gotthard, G, Tanzer, T, Johnsson, K, Hiblot, J. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of the Self-Labeling Protein Tags HaloTag7, SNAP-tag, and CLIP-tag.
Biochemistry, 60, 2021
|
|
1F9N
 
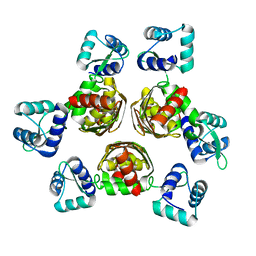 | | CRYSTAL STRUCTURE OF AHRC, THE ARGININE REPRESSOR/ACTIVATOR PROTEIN FROM BACILLUS SUBTILIS | | Descriptor: | ARGININE REPRESSOR/ACTIVATOR PROTEIN | | Authors: | Dennis, C.A, Glykos, N.M, Parsons, M.R, Phillips, S.E.V. | | Deposit date: | 2000-07-11 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of AhrC, the arginine repressor/activator protein from Bacillus subtilis.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
2W24
 
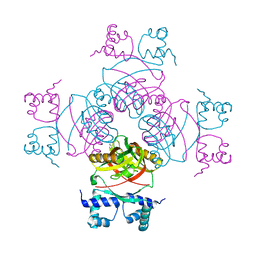 | | M. tuberculosis Rv3291c complexed to Lysine | | Descriptor: | LYSINE, PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, Ramachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
2W25
 
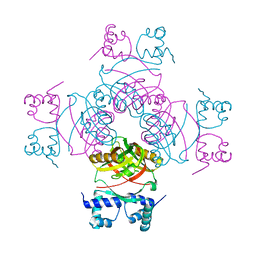 | | Crystal structure of Glu104Ala mutant | | Descriptor: | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN | | Authors: | Shrivastava, T, RAmachandran, R. | | Deposit date: | 2008-10-24 | | Release date: | 2009-11-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Ligand-Induced Structural Transitions, Mutational Analysis, and 'Open' Quaternary Structure of the M. Tuberculosis Feast/Famine Regulatory Protein (Rv3291C).
J.Mol.Biol., 392, 2009
|
|
5V7M
 
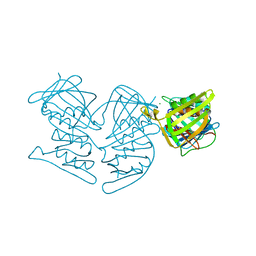 | | PCNA mutant L126A/I128A Protein Defective in Gene Silencing | | Descriptor: | MAGNESIUM ION, Proliferating cell nuclear antigen | | Authors: | Kondratick, C.M, Litman, J.M, Washington, M.T, Dieckman, L.M. | | Deposit date: | 2017-03-20 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of PCNA mutant proteins defective in gene silencing suggest a novel interaction site on the front face of the PCNA ring.
PLoS ONE, 13, 2018
|
|
7L20
 
 | |
3BRJ
 
 | | Crystal structure of mannitol operon repressor (MtlR) from Vibrio parahaemolyticus RIMD 2210633 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Mannitol operon repressor | | Authors: | Tan, K, Zhou, M, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-21 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | The mannitol operon repressor MtlR belongs to a new class of transcription regulators in bacteria.
J.Biol.Chem., 284, 2009
|
|
5V7L
 
 | |
1RR7
 
 | |
2AU0
 
 | | Unmodified preinsertion binary complex | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*G*CP*TP*AP*CP*CP*AP*TP*CP*CP*AP*AP*CP*C)-3', 5'-D(*GP*GP*TP*TP*GP*GP*AP*TP*GP*GP*TP*AP*(DDG))-3', CALCIUM ION, ... | | Authors: | Rechkoblit, O, Malinina, L, Cheng, Y, Kuryavyi, V, Broyde, S, Geacintov, N.E, Patel, D.J. | | Deposit date: | 2005-08-26 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Stepwise Translocation of Dpo4 Polymerase during Error-Free Bypass of an oxoG Lesion
Plos Biol., 4, 2006
|
|
5V3O
 
 | | Cereblon in complex with DDB1 and CC-220 | | Descriptor: | (3S)-3-[4-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1-oxo-1,3-dihydro-2H-isoindol-2-yl]piperidine-2,6-dione, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Matyskiela, M, Pagarigan, B, Chamberlain, P. | | Deposit date: | 2017-03-07 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A Cereblon Modulator (CC-220) with Improved Degradation of Ikaros and Aiolos.
J. Med. Chem., 61, 2018
|
|
5V7K
 
 | |
5UOQ
 
 | | CRYSTAL STRUCTURE OF THE PROTOTYPE FOAMY VIRUS INTASOME WITH A 2- PYRIDINONE AMINAL INHIBITOR (COMPOUND 31) | | Descriptor: | (3R)-8-[(3-chloro-4-fluorophenyl)methyl]-6-hydroxy-1,5,7-trioxo-1,2',3',5,7,8,9,10-octahydro-2H-spiro[imidazo[5,1-a][2,6]naphthyridine-3,1'-indene]-7'-carbonitrile, GLYCEROL, INTEGRASE, ... | | Authors: | Klein, D.J. | | Deposit date: | 2017-02-01 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Discovery and optimization of 2-pyridinone aminal integrase strand transfer inhibitors for the treatment of HIV.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
