8UAB
 
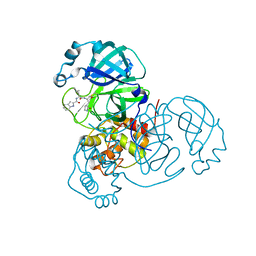 | | SARS-CoV-2 main protease (Mpro) complex with AC1115 | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | DuPrez, K.T, Chao, A, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.781 Å) | | Cite: | SARS-CoV-2 main protease (Mpro) complex with AC1115
To Be Published
|
|
5TNU
 
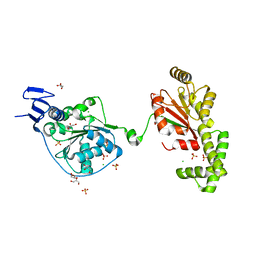 | | S. tokodaii XPB II crystal structure at 3.0 Angstrom resolution | | Descriptor: | CHLORIDE ION, DNA-dependent ATPase XPBII, GLYCEROL, ... | | Authors: | DuPrez, K.T, Hilario, E, Wang, I, Fan, L. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Application of Electrochemical Devices to Characterize the Dynamic Actions of Helicases on DNA.
Anal.Chem., 90, 2018
|
|
5D8N
 
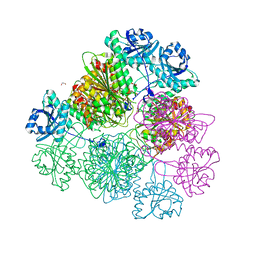 | | Tomato leucine aminopeptidase mutant - K354E | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | DuPrez, K.T, Scranton, M.A, Walling, L.L, Fan, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-05-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural insights into chaperone-activity enhancement by a K354E mutation in tomato acidic leucine aminopeptidase.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
4KSI
 
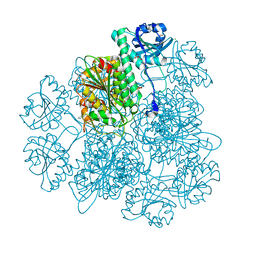 | | Crystal Structure Analysis of the Acidic Leucine Aminopeptidase of Tomato | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | DuPrez, K.T, Scranton, M, Walling, L, Fan, L. | | Deposit date: | 2013-05-17 | | Release date: | 2013-06-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of tomato wound-induced leucine aminopeptidase sheds light on substrate specificity.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6P66
 
 | |
8UAC
 
 | | CATHEPSIN L IN COMPLEX WITH AC1115 | | Descriptor: | Cathepsin L, N-[(2S)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Chao, A, DuPrez, K.T, Han, F.Q. | | Deposit date: | 2023-09-20 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | CATHEPSIN L IN COMPLEX WITH AC1115
To be Published
|
|
6P4W
 
 | |
6P4O
 
 | |
5F4H
 
 | | Archael RuvB-like Holiday junction helicase | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Nucleotide binding protein PINc | | Authors: | Zhai, B, DuPrez, K.T, Doukov, T.I, Shen, Y, Fan, L. | | Deposit date: | 2015-12-03 | | Release date: | 2016-12-21 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Structure and Function of a Novel ATPase that Interacts with Holliday Junction Resolvase Hjc and Promotes Branch Migration.
J. Mol. Biol., 429, 2017
|
|
