1LQ9
 
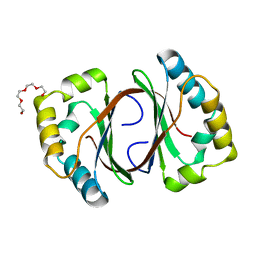 | | Crystal Structure of a Monooxygenase from the Gene ActVA-Orf6 of Streptomyces coelicolor Strain A3(2) | | Descriptor: | ACTVA-ORF6 MONOOXYGENASE, TETRAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-05-09 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
EMBO J., 22, 2003
|
|
5L43
 
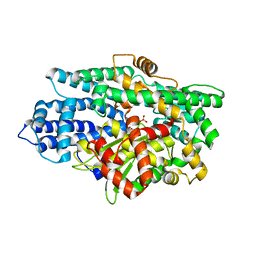 | | Structure of K26-DCP | | Descriptor: | K-26 dipeptidyl carboxypeptidase, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Masuyer, G, Acharya, K.R, Kramer, G.J, Bachmann, B.O. | | Deposit date: | 2016-05-24 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a peptidyl-dipeptidase K-26-DCP from Actinomycete in complex with its natural inhibitor.
FEBS J., 283, 2016
|
|
5LMZ
 
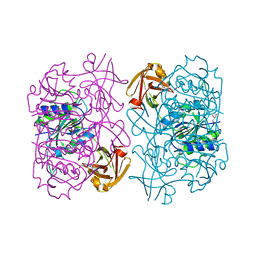 | | Fluorinase from Streptomyces sp. MA37 | | Descriptor: | 1-DEAZA-ADENOSINE, CHLORIDE ION, Fluorinase, ... | | Authors: | Mann, G, O'Hagan, D, Naismith, J.H, Bandaranayaka, N. | | Deposit date: | 2016-08-02 | | Release date: | 2016-08-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification of fluorinases from Streptomyces sp MA37, Norcardia brasiliensis, and Actinoplanes sp N902-109 by genome mining.
Chembiochem, 15, 2014
|
|
6S8Z
 
 | |
3JXG
 
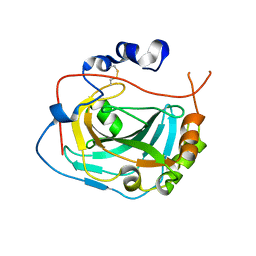 | | CA-like domain of mouse PTPRG | | Descriptor: | Receptor-type tyrosine-protein phosphatase gamma | | Authors: | Bouyain, S. | | Deposit date: | 2009-09-19 | | Release date: | 2009-12-22 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The protein tyrosine phosphatases PTPRZ and PTPRG bind to distinct members of the contactin family of neural recognition molecules.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1GWY
 
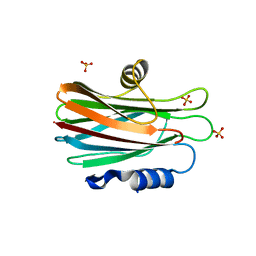 | | Crystal structure of the water-soluble state of the pore-forming cytolysin Sticholysin II | | Descriptor: | STICHOLYSIN II, SULFATE ION | | Authors: | Mancheno, J.M, Martin-Benito, J, Martinez-Ripoll, M, Gavilanes, J.G, Hermoso, J.A. | | Deposit date: | 2002-03-26 | | Release date: | 2003-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal and Electron Microscopy Structures of Sticholysin II Actinoporin Reveal Insights Into the Mechanism of Membrane Pore Formation
Structure, 11, 2003
|
|
7X6R
 
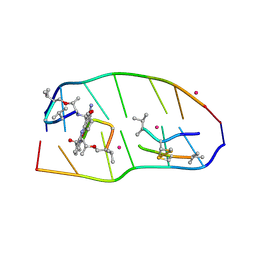 | | Crystal structure of actinomycin D-echinomycin-d(AGCACGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, COBALT (II) ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-08 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
7X97
 
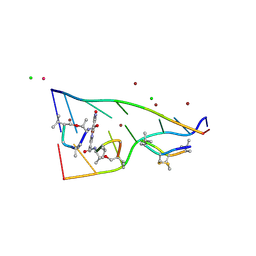 | | Crystal structure of actinomycin D-echinomycin-d(AGCCCGT/ACGGGCT) complex | | Descriptor: | 2-CARBOXYQUINOXALINE, Actinomycin D, CHLORIDE ION, ... | | Authors: | Kao, S.H, Satange, R.B, Hou, M.H. | | Deposit date: | 2022-03-15 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Staggered intercalation of DNA duplexes with base-pair modulation by two distinct drug molecules induces asymmetric backbone twisting and structure polymorphism.
Nucleic Acids Res., 50, 2022
|
|
1N5Q
 
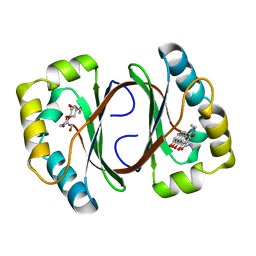 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with dehydrated Sancycline | | Descriptor: | 4-DIMETHYLAMINO-1,10,11,12-TETRAHYDROXY-3-OXO-3,4,4A,5-TETRAHYDRO-NAPHTHACENE-2-CARBOXYLIC ACID AMIDE, ActaVA-Orf6 monooxygenase, HEXAETHYLENE GLYCOL | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5S
 
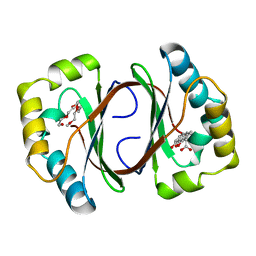 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9-OXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5T
 
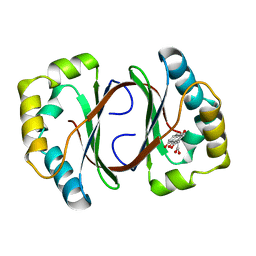 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Oxidized Acetyl Dithranol | | Descriptor: | (1,8-DIHYDROXY-9,10-DIOXO-9,10-DIHYDRO-ANTHRACEN-2-YL)-ACETIC ACID, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, G Kendrew, S, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
1N5V
 
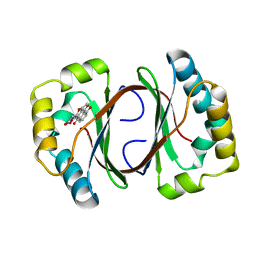 | | Crystal structure of a Monooxygenase from the gene ActVA-Orf6 of Streptomyces coelicolor in complex with the ligand Nanaomycin D | | Descriptor: | 7-HYDROXY-5-METHYL-3,3A,5,11B-TETRAHYDRO-1,4-DIOXA-CYCLOPENTA[A]ANTHRACENE-2,6,11-TRIONE, ActVA-Orf6 monooxygenase | | Authors: | Sciara, G, Kendrew, S.G, Miele, A.E, Marsh, N.G, Federici, L, Malatesta, F, Schimperna, G, Savino, C, Vallone, B. | | Deposit date: | 2002-11-07 | | Release date: | 2003-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The structure of ActVA-Orf6, a novel type of monooxygenase involved in
actinorhodin biosynthesis
Embo J., 22, 2003
|
|
2HL5
 
 | |
1RIR
 
 | | Crystal structure of meso-tetrasulphonatophenylporphyrin in complex with Peanut lectin. | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Goel, M, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-11-17 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the PNA-porphyrin complex in the presence and absence of lactose: mapping the conformational changes on lactose binding, interacting surfaces, and supramolecular aggregations.
Biochemistry, 44, 2005
|
|
1QR0
 
 | | CRYSTAL STRUCTURE OF THE 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP-COENZYME A COMPLEX | | Descriptor: | 4'-PHOSPHOPANTETHEINYL TRANSFERASE SFP, COENZYME A, MAGNESIUM ION | | Authors: | Reuter, K, Mofid, R.M, Marahiel, A.M, Ficner, R. | | Deposit date: | 1999-06-17 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the surfactin synthetase-activating enzyme sfp: a prototype of the 4'-phosphopantetheinyl transferase superfamily.
EMBO J., 18, 1999
|
|
2XUZ
 
 | | Crystal structure of the triscatecholate siderophore binding protein FeuA from Bacillus subtilis complexed with Ferri-Enterobactin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, IRON-UPTAKE SYSTEM-BINDING PROTEIN, ... | | Authors: | Peuckert, F, Miethke, M, Schwoerer, C.J, Albrecht, A.G, Oberthuer, M, Marahiel, M.A. | | Deposit date: | 2010-10-22 | | Release date: | 2011-08-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Siderophore Binding Protein Feua Shows Limited Promiscuity Toward Exogenous Triscatecholates
Chem.Biol., 18, 2011
|
|
1RIT
 
 | | Crystal structure of Peanut lectin in complex with meso-tetrasulphonatophenylporphyrin and lactose | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, CALCIUM ION, Galactose-binding lectin, ... | | Authors: | Goel, M, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-11-17 | | Release date: | 2004-12-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structures of the PNA-porphyrin complex in the presence and absence of lactose: mapping the conformational changes on lactose binding, interacting surfaces, and supramolecular aggregations.
Biochemistry, 44, 2005
|
|
2IOU
 
 | |
4TMC
 
 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
6EPZ
 
 | |
6EQ0
 
 | |
6EQ1
 
 | |
6EPY
 
 | |
2RHR
 
 | | P94L actinorhodin ketordeuctase mutant, with NADPH and Inhibitor Emodin | | Descriptor: | 3-METHYL-1,6,8-TRIHYDROXYANTHRAQUINONE, Actinorhodin Polyketide Ketoreductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Korman, T.P, Tsai, S.-C. | | Deposit date: | 2007-10-09 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase
Biochemistry, 47, 2008
|
|
2Y3M
 
 | | Structure of the extra-membranous domain of the secretin HofQ from Actinobacillus actinomycetemcomitans | | Descriptor: | FORMIC ACID, GLYCEROL, PROTEIN TRANSPORT PROTEIN HOFQ | | Authors: | Tarry, M, Jaaskelainen, M, Tuominen, H, Ihalin, R, Hogbom, M. | | Deposit date: | 2010-12-21 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Extra-Membranous Domains of the Competence Protein Hofq Show DNA Binding, Flexibility and a Shared Fold with Type I Kh Domains.
J.Mol.Biol., 409, 2011
|
|
