4F2J
 
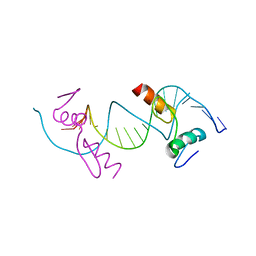 | | Crystal structure of ZNF217 bound to DNA, P6522 crystal form | | Descriptor: | 5'-D(*TP*TP*TP*GP*CP*AP*GP*AP*AP*TP*CP*GP*AP*TP*TP*CP*TP*GP*CP*A)-3', ZINC ION, Zinc finger protein 217 | | Authors: | Vandevenne, M.S, Jacques, D.A, Guss, J.M, Mackay, J.P. | | Deposit date: | 2012-05-08 | | Release date: | 2013-02-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | New insights into DNA recognition by zinc fingers revealed by structural analysis of the oncoprotein ZNF217
J.Biol.Chem., 288, 2013
|
|
3WFX
 
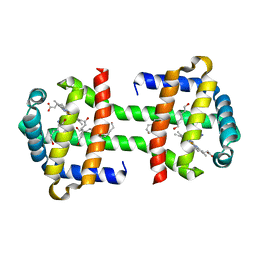 | | Crystal Structure of the Imidazole-Bound Form of the HGbRL's Globin Domain | | Descriptor: | HEXANE-1,6-DIOL, Hemoglobin-like flavoprotein fused to Roadblock/LC7 domain, IMIDAZOLE, ... | | Authors: | Teh, A.H. | | Deposit date: | 2013-07-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Open and Lys-His Hexacoordinated Closed Structures of a Globin with Swapped Proximal and Distal Sites.
Sci Rep, 5, 2015
|
|
4J62
 
 | |
4J69
 
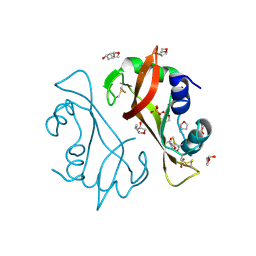 | | Crystal structure of Ribonuclease A soaked in 50% S,R,S-bisfuranol: One of twelve in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
4J6N
 
 | |
3WEC
 
 | | Structure of P450 RauA (CYP1050A1) complexed with a biosynthetic intermediate of aurachin RE | | Descriptor: | 3-[(2E,6E,9R)-9-hydroxy-3,7,11-trimethyldodeca-2,6,10-trien-1-yl]-2-methylquinolin-4(1H)-one, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Yasutake, Y, Kitagawa, W, Tamura, T. | | Deposit date: | 2013-07-03 | | Release date: | 2014-01-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of the quinoline N-hydroxylating cytochrome P450 RauA, an essential enzyme that confers antibiotic activity on aurachin alkaloids
Febs Lett., 588, 2014
|
|
3WHM
 
 | | Structure of Hemoglobin Complex with 18-crown-6 | | Descriptor: | 1,4,7,10,13,16-HEXAOXACYCLOOCTADECANE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Lee, C.C, Lin, L.L, Wang, A.H.J. | | Deposit date: | 2013-08-27 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crowning proteins: modulating the protein surface properties using crown ethers.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3WGE
 
 | | Crystal structure of ERp46 Trx2 | | Descriptor: | GLYCEROL, Thioredoxin domain-containing protein 5 | | Authors: | Inaba, K, Suzuki, M, Kojima, R. | | Deposit date: | 2013-08-04 | | Release date: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Radically different thioredoxin domain arrangement of ERp46, an efficient disulfide bond introducer of the mammalian PDI family
Structure, 22, 2014
|
|
3EXO
 
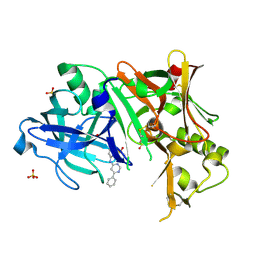 | | Crystal structure of BACE1 bound to inhibitor | | Descriptor: | Beta-secretase 1, GLYCEROL, N-{2-methyl-5-[(6-phenylpyrimidin-4-yl)amino]phenyl}methanesulfonamide, ... | | Authors: | Allison, T.J. | | Deposit date: | 2008-10-16 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of a small molecule beta-secretase inhibitor that binds without catalytic aspartate engagement.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EXX
 
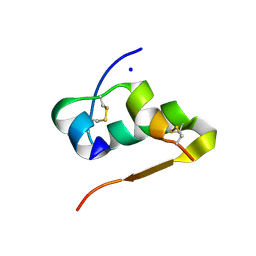 | |
3WIU
 
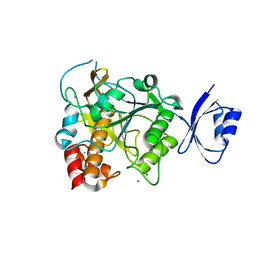 | |
3NC3
 
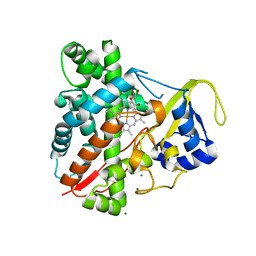 | | CYP134A1 structure with a closed substrate binding loop | | Descriptor: | Cytochrome P450 cypX, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cryle, M.J, Schlichting, I. | | Deposit date: | 2010-06-04 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structural and biochemical characterization of the cytochrome P450 CypX (CYP134A1) from Bacillus subtilis: a cyclo-L-leucyl-L-leucyl dipeptide oxidase.
Biochemistry, 49, 2010
|
|
4DXY
 
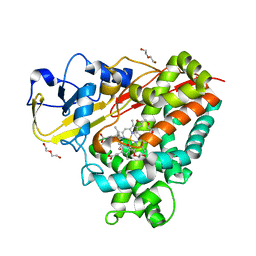 | | Crystal structures of CYP101D2 Y96A mutant | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zhou, W, Bell, S.G, Yang, W, Dale, A, Wong, L.-L. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improving the affinity and activity of CYP101D2 for hydrophobic substrates
Appl.Microbiol.Biotechnol., 2012
|
|
4I0D
 
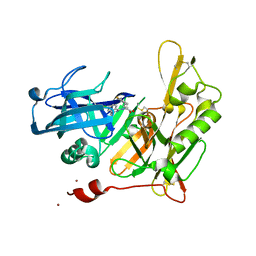 | |
3WMK
 
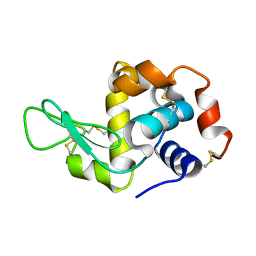 | |
4DY9
 
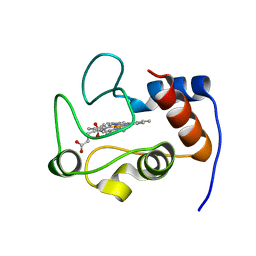 | |
3F8A
 
 | | Crystal Structure of the R132K:R111L:L121E:R59W Mutant of Cellular Retinoic Acid-Binding Protein Type II Complexed with C15-aldehyde (a retinal analog) at 1.95 Angstrom resolution. | | Descriptor: | 1,3,3-trimethyl-2-[(1E,3E)-3-methylpenta-1,3-dien-1-yl]cyclohexene, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-12 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Probing Wavelength Regulation with an Engineered Rhodopsin Mimic and a C15-Retinal Analogue
Chempluschem, 77, 2012
|
|
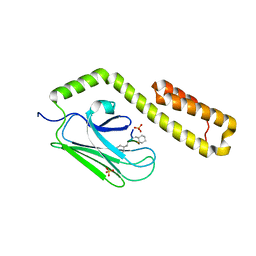
4HY9
 
 | |
3PLB
 
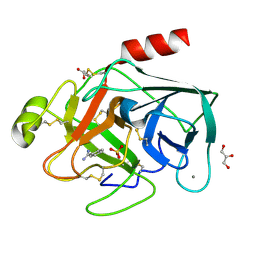 | | Bovine trypsin variant X(tripleIle227) in complex with small molecule inhibitor | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4DTW
 
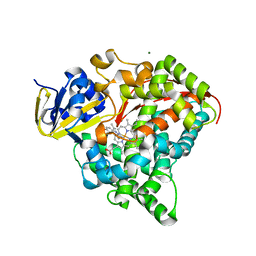 | | cytochrome P450 BM3h-8C8 MRI sensor bound to serotonin | | Descriptor: | Cytochrome P450 BM3 variant 8C8, MAGNESIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
3PLP
 
 | | Bovine trypsin variant X(tripleIle227) in complex with small molecule inhibitor | | Descriptor: | 2,7-BIS-(4-AMIDINOBENZYLIDENE)-CYCLOHEPTAN-1-ONE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Braeuer, U, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-11-15 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
3WRK
 
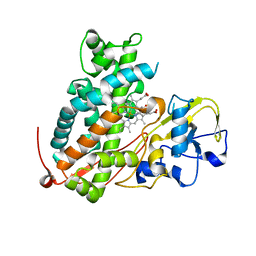 | | Crystal structure of P450cam | | Descriptor: | CAMPHOR, Camphor 5-monooxygenase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kishimoto, A, Takagi, K, Amano, A, Sakurai, K, Mizushima, T, Shimada, H. | | Deposit date: | 2014-02-25 | | Release date: | 2015-03-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure of P450cam intermedite
To be Published
|
|
4I3C
 
 | | Crystal structure of fluorescent protein UnaG N57Q mutant | | Descriptor: | 3-[5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-2-[[5-[(Z)-(3-ethenyl-4-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, Bilirubin-inducible fluorescent protein UnaG | | Authors: | Kumagai, A, Ando, R, Miyatake, H, Miyawaki, A. | | Deposit date: | 2012-11-26 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | A bilirubin-inducible fluorescent protein from eel muscle
Cell(Cambridge,Mass.), 153, 2013
|
|
4DUA
 
 | | cytochrome P450 BM3h-9D7 MRI sensor, no ligand | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRIETHYLENE GLYCOL, cytochrome P450 BM3 variant 9D7 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
3PNA
 
 | | Crystal Structure of cAMP bound (91-244)RIa Subunit of cAMP-dependent Protein Kinase | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, GLYCEROL, cAMP-dependent protein kinase type I-alpha regulatory subunit | | Authors: | Kim, C, Taylor, S. | | Deposit date: | 2010-11-18 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.503 Å) | | Cite: | Cyclic AMP Analog Blocks Kinase Activation by Stabilizing Inactive Conformation: Conformational Selection Highlights a New Concept in Allosteric Inhibitor Design.
Mol Cell Proteomics, 10, 2011
|
|
