4D2Y
 
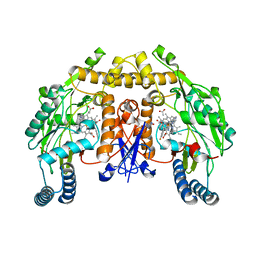 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with (1R,2R)-2-(3-fluorobenzyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}cyclopropanamine | | Descriptor: | (1R,2R)-2-(3-fluorobenzyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}cyclopropanamine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
6MPF
 
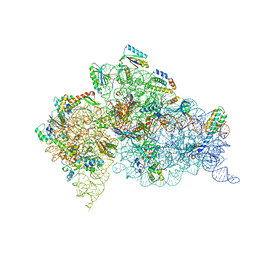 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a 2-thiocytidine (s2C32) and inosine (I34) modified anticodon stem loop (ASL) of Escherichia coli transfer RNA Arginine 1 (TRNAARG1) bound to an mRNA with an CGC-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Cantara, W.A, DeMirci, H, Agris, P.F. | | Deposit date: | 2018-10-05 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | A Structural Basis for Restricted Codon Recognition Mediated by 2-thiocytidine in tRNA Containing a Wobble Position Inosine.
J.Mol.Biol., 432, 2020
|
|
2A8W
 
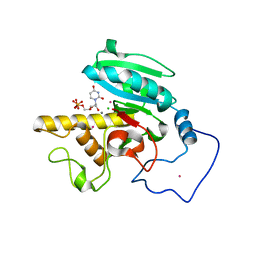 | | Crystal Structure of Human N-acetylgalactosaminyltransferase (GTA) Complexed with Beta-Methyllactoside | | Descriptor: | CHLORIDE ION, Histo-blood group ABO system transferase (NAGAT) [Includes: Glycoprotein-fucosylgalactoside alpha-N- acetylgalactosaminyltransferase (EC 2.4.1.40) (Fucosylglycoprotein alpha-N-acetylgalactosaminyltransferase) (Histo-blood group A transferase) (A transferase); Glycoprotein-fucosylgalactoside alpha- galactosyltransferase (EC 2.4.1.37) (Fucosylglycoprotein 3-alpha- galactosyltransferase) (Histo-blood group B transferase) (B transferase)] [Contains: Fucosylglycoprotein alpha-N- acetylgalactosaminyltransferase, soluble form], ... | | Authors: | Letts, J.A, Rose, N.L, Fang, Y.R, Barry, C.H, Borisova, S.N, Seto, N.O, Palcic, M.M, Evans, S.V. | | Deposit date: | 2005-07-09 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Differential Recognition of the Type I and II H Antigen Acceptors by the Human ABO(H) Blood Group A and B Glycosyltransferases.
J.Biol.Chem., 281, 2006
|
|
6VJN
 
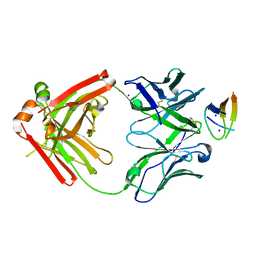 | | Structure of NHP D11A.B5Fab in complex with 16055 V2b peptide | | Descriptor: | D11A.B5 Fab Heavy chain, D11A.B5 Fab Light chain, SODIUM ION, ... | | Authors: | Singh, S, Liban, T.J, Pancera, M. | | Deposit date: | 2020-01-16 | | Release date: | 2021-01-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structurally related but genetically unrelated antibody lineages converge on an immunodominant HIV-1 Env neutralizing determinant following trimer immunization.
Plos Pathog., 17, 2021
|
|
4D31
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)-N-(3-cyanobenzyl) ethan-1-amine | | Descriptor: | 3-[({2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}amino)methyl]benzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
6VM5
 
 | | Structure of Moraxella osloensis Cap4 SAVED/CARF-domain containing receptor | | Descriptor: | MAGNESIUM ION, SAVED domain-containing protein | | Authors: | Lowey, B, Whiteley, A.T, Keszei, A.F.A, Morehouse, B.R, Antine, S.P, Cabrera, V, Schwede, F, Mekalanos, J.J, Shao, S, Lee, A.S.Y, Kranzusch, P.J. | | Deposit date: | 2020-01-27 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | CBASS Immunity Uses CARF-Related Effectors to Sense 3'-5'- and 2'-5'-Linked Cyclic Oligonucleotide Signals and Protect Bacteria from Phage Infection.
Cell, 182, 2020
|
|
6VMV
 
 | | Crystal structure of the H767A mutant of GoxA soaked with glycine | | Descriptor: | Glycine oxidase, MAGNESIUM ION, SULFATE ION | | Authors: | Yukl, E.T. | | Deposit date: | 2020-01-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Roles of active-site residues in catalysis, substrate binding, cooperativity, and the reaction mechanism of the quinoprotein glycine oxidase.
J.Biol.Chem., 295, 2020
|
|
2L74
 
 | | Solution structure of the PilZ domain protein PA4608 complex with c-di-GMP identifies charge clustering as molecular readout | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Putative uncharacterized protein PA4608 | | Authors: | Habazettl, J, Allan, M, Jenal, U, Grzesiek, S. | | Deposit date: | 2010-12-02 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the PilZ domain protein PA4608 complex with cyclic di-GMP identifies charge clustering as molecular readout
J.Biol.Chem., 286, 2011
|
|
4D8J
 
 | | Structure of E. coli MatP-mats complex | | Descriptor: | 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*AP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', 5'-D(*TP*TP*CP*GP*TP*GP*AP*CP*AP*TP*TP*GP*TP*CP*AP*CP*GP*AP*A)-3', Macrodomain Ter protein | | Authors: | Dupaigne, P, Tonthat, N.K, Espeli, O, Whitfill, T, Boccard, F, Schumacher, M.A. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Molecular basis for a protein-mediated DNA-bridging mechanism that functions in condensation of the E. coli chromosome.
Mol.Cell, 48, 2012
|
|
6VMK
 
 | |
4D9O
 
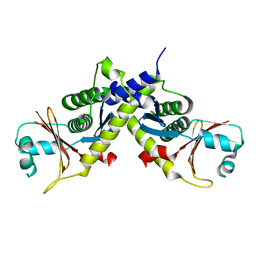 | | Structure of ebolavirus protein VP24 from Reston | | Descriptor: | Membrane-associated protein VP24 | | Authors: | Zhang, A.P.P. | | Deposit date: | 2012-01-11 | | Release date: | 2012-04-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The ebola virus interferon antagonist VP24 directly binds STAT1 and has a novel, pyramidal fold
Plos Pathog., 8, 2012
|
|
4D9W
 
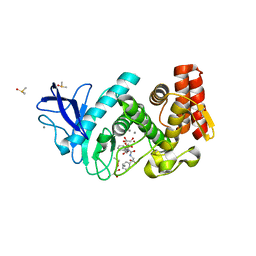 | |
6VOK
 
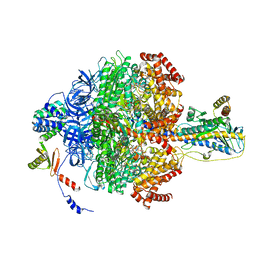 | | Chloroplast ATP synthase (R3, CF1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase delta chain, ... | | Authors: | Yang, J.-H, Williams, D, Kandiah, E, Fromme, P, Chiu, P.-L. | | Deposit date: | 2020-01-30 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis of redox modulation on chloroplast ATP synthase.
Commun Biol, 3, 2020
|
|
2LNW
 
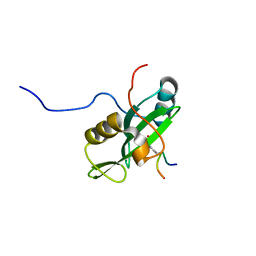 | | Identification and structural basis for a novel interaction between Vav2 and Arap3 | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3, Guanine nucleotide exchange factor VAV2 | | Authors: | Wu, B, Zhang, J, Wu, J, Shi, Y. | | Deposit date: | 2012-01-05 | | Release date: | 2012-11-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and structural basis for a novel interaction between Vav2 and Arap3.
J.Struct.Biol., 180, 2012
|
|
6N57
 
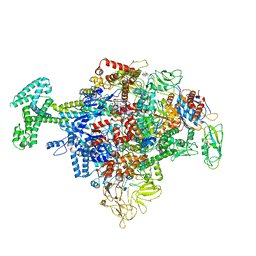 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound with TraR in conformation I | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2018-11-21 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6VQ9
 
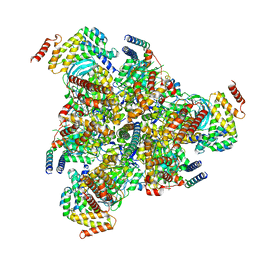 | |
4CWX
 
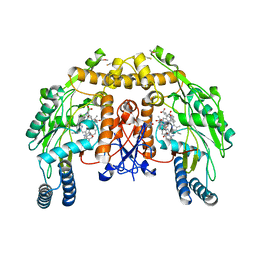 | | ALPHA-KETOGLUTARATE-DEPENDENT DIOXYGENASE COMPLEX1 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[(3R,4R)-4-{[5-(6-aminopyridin-2-yl)pentyl]oxy}pyrrolidin-3-yl]methyl}-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-04-03 | | Release date: | 2014-08-13 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mobility of a Conserved Tyrosine Residue Controls Isoform-Dependent Enzyme-Inhibitor Interactions in Nitric Oxide Synthases.
Biochemistry, 53, 2014
|
|
6MH8
 
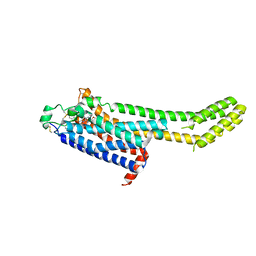 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a, Soluble cytochrome b562 chimeric construct | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
6VVF
 
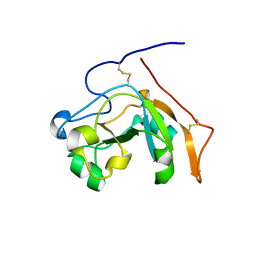 | |
4D4F
 
 | | Mutant P250A of bacterial chalcone isomerase from Eubacterium ramulus | | Descriptor: | CHALCONE ISOMERASE, CHLORIDE ION, GLYCEROL | | Authors: | Thomsen, M, Kratzat, H, Hinrichs, W. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for (2 R ,3 R )-Taxifolin Binding and Reaction Products to the Bacterial Chalcone Isomerase of Eubacterium ramulus.
Molecules, 27, 2022
|
|
4MH2
 
 | | Crystal structure of Bovine Mitochondrial Peroxiredoxin III | | Descriptor: | CITRIC ACID, Thioredoxin-dependent peroxide reductase, mitochondrial | | Authors: | Cao, Z, McGow, D.P, Shepherd, C, Lindsay, J.G. | | Deposit date: | 2013-08-29 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Improved Catenated Structures of Bovine Peroxiredoxin III F190L Reveal Details of Ring-Ring Interactions and a Novel Conformational State.
Plos One, 10, 2015
|
|
2A1T
 
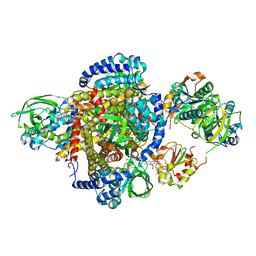 | | Structure of the human MCAD:ETF E165betaA complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Acyl-CoA dehydrogenase, medium-chain specific, ... | | Authors: | Toogood, H.S, Van Thiel, A, Scrutton, N.S, Leys, D. | | Deposit date: | 2005-06-21 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stabilization of Non-productive Conformations Underpins Rapid Electron Transfer to Electron-transferring Flavoprotein
J.Biol.Chem., 280, 2005
|
|
4CID
 
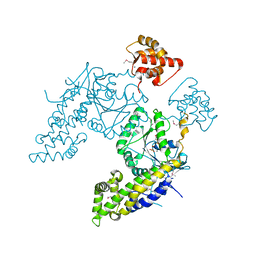 | |
6VZA
 
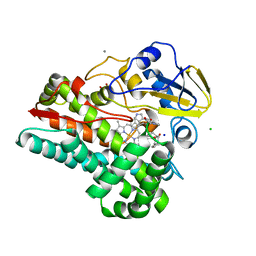 | | Crystal structure of cytochrome P450 NasF5053 Q65I-A86G mutant variant from Streptomyces sp. NRRL F-5053 in the cyclo-L-Trp-L-Pro-bound state | | Descriptor: | (3S,8aS)-3-(1H-indol-3-ylmethyl)hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Luo, Z, Jia, X, Sun, C, Qu, X, Kobe, B. | | Deposit date: | 2020-02-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular basis of regio- and stereo-specificity in biosynthesis of bacterial heterodimeric diketopiperazines.
Nat Commun, 11, 2020
|
|
2L6J
 
 | | Tah1 complexed by MEEVD | | Descriptor: | C-terminus Hsp90 chaperone peptide MEEVD, TPR repeat-containing protein associated with Hsp90 | | Authors: | Jimenez, B, Ugwu, F, Zhao, R, Orti, L, Houry, W.A, Pineda-Lucena, A. | | Deposit date: | 2010-11-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of minimal tetratricopeptide repeat domain protein Tah1 reveals mechanism of its interaction with Pih1 and Hsp90.
J.Biol.Chem., 287, 2012
|
|
