3CBS
 
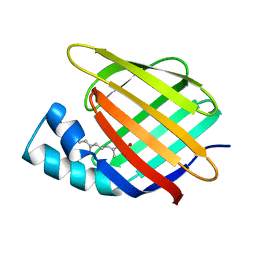 | | CELLULAR RETINOIC ACID BINDING PROTEIN II IN COMPLEX WITH A SYNTHETIC RETINOIC ACID (RO-12 7310) | | Descriptor: | (2E,4E,6E,8E)-9-(4-hydroxy-2,3,6-trimethylphenyl)-3,7-dimethylnona-2,4,6,8-tetraenoic acid, PROTEIN (CRABP-II) | | Authors: | Chaudhuri, B, Kleywegt, G.J, Bergfors, T, Jones, T.A. | | Deposit date: | 1999-02-22 | | Release date: | 1999-12-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of cellular retinoic acid binding proteins I and II in complex with synthetic retinoids.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1GAE
 
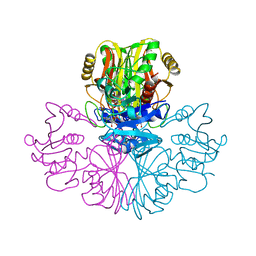 | | COMPARISON OF THE STRUCTURES OF WILD TYPE AND A N313T MUTANT OF ESCHERICHIA COLI GLYCERALDEHYDE 3-PHOSPHATE DEHYDROGENASES: IMPLICATION FOR NAD BINDING AND COOPERATIVITY | | Descriptor: | D-GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Duee, E, Olivier-Deyris, L, Fanchon, E, Corbier, C, Branlant, G, Dideberg, O. | | Deposit date: | 1995-10-24 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity.
J.Mol.Biol., 257, 1996
|
|
2R2M
 
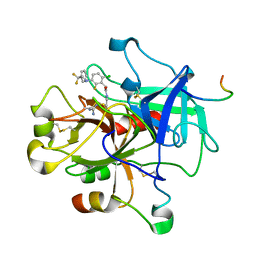 | | 2-(2-Chloro-6-Fluorophenyl)Acetamides as Potent Thrombin Inhibitors | | Descriptor: | Hirudin-3A, N-[2-({[amino(imino)methyl]amino}oxy)ethyl]-2-{6-chloro-3-[(2,2-difluoro-2-phenylethyl)amino]-2-fluorophenyl}acetamide, Thrombin heavy chain, ... | | Authors: | Spurlino, J. | | Deposit date: | 2007-08-27 | | Release date: | 2008-08-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | 2-(2-Chloro-6-Fluorophenyl)Acetamides as Potent Thrombin Inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1AM7
 
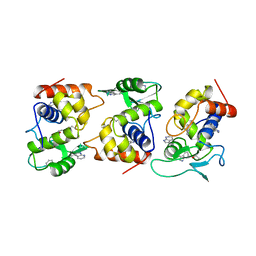 | | Lysozyme from bacteriophage lambda | | Descriptor: | ISOPROPYL ALCOHOL, LYSOZYME | | Authors: | Evrard, C, Fastrez, J, Declercq, J.P. | | Deposit date: | 1997-06-24 | | Release date: | 1997-12-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the lysozyme from bacteriophage lambda and its relationship with V and C-type lysozymes.
J.Mol.Biol., 276, 1998
|
|
315D
 
 | |
3AAJ
 
 | | Crystal structure of Ca2+-bound form of des3-23ALG-2deltaGF122 | | Descriptor: | CALCIUM ION, Programmed cell death protein 6 | | Authors: | Suzuki, H, Inuzuka, T, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
3KUH
 
 | | Crystal structure of E. coli HPPK(H115A) in complex with AMPCPP and HP | | Descriptor: | 2-AMINO-6-HYDROXYMETHYL-7,8-DIHYDRO-3H-PTERIDIN-4-ONE, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase, ACETATE ION, ... | | Authors: | Blaszczyk, J, Li, Y, Yan, H, Ji, X. | | Deposit date: | 2009-11-27 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Roles of residues E77 and H115 in E. coli HPPK
To be Published
|
|
3CI2
 
 | |
3UB5
 
 | |
3AAK
 
 | | Crystal structure of Zn2+-bound form of des3-20ALG-2F122A | | Descriptor: | Programmed cell death protein 6, ZINC ION | | Authors: | Inuzuka, T, Suzuki, H, Kawasaki, M, Shibata, H, Wakatsuki, S, Maki, M. | | Deposit date: | 2009-11-19 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for defect in Alix-binding by alternatively spliced isoform of ALG-2 (ALG-2DeltaGF122) and structural roles of F122 in target recognition
Bmc Struct.Biol., 10, 2010
|
|
2GI4
 
 | |
3LUH
 
 | |
3LUG
 
 | |
3U4L
 
 | |
3CYT
 
 | |
2LVB
 
 | | Solution NMR Structure DE NOVO DESIGNED PFK fold PROTEIN, Northeast Structural Genomics Consortium (NESG) Target OR250 | | Descriptor: | DE NOVO DESIGNED PFK fold PROTEIN | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-30 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
3CEL
 
 | | ACTIVE-SITE MUTANT E212Q DETERMINED AT PH 6.0 WITH CELLOBIOSE BOUND IN THE ACTIVE SITE | | Descriptor: | 1,4-BETA-D-GLUCAN CELLOBIOHYDROLASE I, 2-acetamido-2-deoxy-beta-D-glucopyranose, CADMIUM ION, ... | | Authors: | Divne, C, Stahlberg, J, Jones, T.A. | | Deposit date: | 1996-08-24 | | Release date: | 1997-03-12 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Activity studies and crystal structures of catalytically deficient mutants of cellobiohydrolase I from Trichoderma reesei.
J.Mol.Biol., 264, 1996
|
|
1HO0
 
 | | NEW B-CHAIN MUTANT OF BOVINE INSULIN | | Descriptor: | INSULIN | | Authors: | Dupradeau, F.Y, Richard, T, Le Flem, G, Oulyadi, H, Prigent, Y, Monti, J.P. | | Deposit date: | 2000-12-08 | | Release date: | 2000-12-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new B-chain mutant of insulin: comparison with the insulin crystal structure and role of sulfonate groups in the B-chain structure
J.Pept.Res., 60, 2002
|
|
2LTA
 
 | | Solution NMR structure of De novo designed protein, rossmann 3x1 fold, Northeast Structural Genomics Consortium target OR157 | | Descriptor: | De novo designed protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Kohan, E, Acton, T.B, Kornhaber, G, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-05-15 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
2R96
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2R97
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
2LV8
 
 | | Solution NMR Structure de novo designed rossmann 2x2 fold protein, Northeast Structural Genomics Consortium (NESG) Target OR16 | | Descriptor: | De novo designed rossmann 2x2 fold protein | | Authors: | Liu, G, Koga, R, Koga, N, Xiao, R, Pederson, K, Hamilton, K, Ciccosanti, C, Acton, T.B, Everett, J.K, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-06-29 | | Release date: | 2012-08-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
9CEX
 
 | | Spizellomyces punctatus Fanzor (SpuFz) State 4 | | Descriptor: | DNA (29-MER), DNA (5'-D(*(MG)*(MG)P*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*AP*A)-3'), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), ... | | Authors: | Xu, P, Saito, M, Zhang, F. | | Deposit date: | 2024-06-27 | | Release date: | 2024-09-11 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into the diversity and DNA cleavage mechanism of Fanzor.
Cell, 187, 2024
|
|
8CBY
 
 | | Crystal Structure of Anti-cortisol Fab in Complex with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol (17) Fab (heavy chain), anti-cortisol (17) Fab (light chain) | | Authors: | Eronen, V, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insight to elucidate the binding specificity of the anti-cortisol Fab fragment with glucocorticoids.
J.Struct.Biol., 215, 2023
|
|
8C4J
 
 | | CdaA-compound 4 complex | | Descriptor: | 5-methylpyrimidin-4-amine, CHLORIDE ION, Diadenylate cyclase | | Authors: | Neumann, P, Ficner, R. | | Deposit date: | 2023-01-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Computer-aided design of a cyclic di-AMP synthesizing enzyme CdaA inhibitor.
Microlife, 4, 2023
|
|
