3U6W
 
 | | Truncated M. tuberculosis LeuA (1-425) complexed with KIV | | Descriptor: | 2-isopropylmalate synthase, 3-METHYL-2-OXOBUTANOIC ACID, GLYCEROL, ... | | Authors: | Koon, N, Baker, E.N, Squire, C.J. | | Deposit date: | 2011-10-13 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Removal of the C-terminal regulatory domain of alpha-isopropylmalate synthase disrupts functional substrate binding.
Biochemistry, 51, 2012
|
|
7K5M
 
 | | CRYSTAL STRUCTURE OF HBV CAPSID Y132A MUTANT IN COMPLEX WITH N-(3-chloro-4-fluorophenyl)-3-phenyl-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide AT 2.65A RESOLUTION | | Descriptor: | Capsid protein, ISOPROPYL ALCOHOL, N-(3-chloro-4-fluorophenyl)-3-phenyl-2,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridine-5-carboxamide | | Authors: | Kuduk, S.D, Stoops, B, Alexander, R, Lam, A.M, Espiritu, C, Vogel, R, Lau, V, Klumpp, K, Flores, O.A, Hartman, G.D, Lukacs, C.M, Abendroth, J. | | Deposit date: | 2020-09-17 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Identification of a new class of HBV capsid assembly modulator.
Bioorg.Med.Chem.Lett., 39, 2021
|
|
2QSR
 
 | | Crystal structure of C-terminal domain of transcription-repair coupling factor | | Descriptor: | Transcription-repair coupling factor | | Authors: | Ramagopal, U.A, Toro, R, Gilmore, M, Bain, K, Iizuka, M, Wasserman, S, Rodgers, L, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-07-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of C-terminal domain of transcription-repair coupling factor.
To be Published
|
|
6T3D
 
 | | Crystal structure of AmpC from E.coli | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Lang, P.A, Leissing, T.M, Schofield, C.J, Brem, J. | | Deposit date: | 2019-10-10 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bicyclic Boronates as Potent Inhibitors of AmpC, the Class C beta-Lactamase from Escherichia coli .
Biomolecules, 10, 2020
|
|
5VRC
 
 | |
2XYU
 
 | | Crystal structure of EphA4 kinase domain in complex with VUF 12058 | | Descriptor: | 5-(5-FLUORO-2-METHYLPHENYL)-6,7,8,9-TETRAHYDRO-3H-PYRAZOLO[3,4-C]ISOQUINOLIN-1-AMINE, EPHRIN TYPE-A RECEPTOR 4,, GLYCEROL | | Authors: | Farenc, C.J.A, Celie, P.H.N, vanLinden, O.P.J, Siegal, G. | | Deposit date: | 2010-11-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Fragment Based Lead Discovery of Small Molecule Inhibitors for the Epha4 Receptor Tyrosine Kinase.
Eur.J.Med.Chem., 47, 2012
|
|
6EFK
 
 | | Crystal structure of the human CHIP TPR domain in complex with a 5mer acetylated HSP70 peptide | | Descriptor: | ACE-ILE-GLU-GLU-VAL-ASP, E3 ubiquitin-protein ligase CHIP, SODIUM ION | | Authors: | Basu, K, Ravalin, M, Bohn, M.-F, Craik, C.S, Gestwicki, J.E. | | Deposit date: | 2018-08-16 | | Release date: | 2019-07-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Specificity for latent C termini links the E3 ubiquitin ligase CHIP to caspases.
Nat.Chem.Biol., 15, 2019
|
|
6ALE
 
 | | A V-to-F substitution in SK2 channels causes Ca2+ hypersensitivity and improves locomotion in a C. elegans ALS model | | Descriptor: | (3E)-6,7-dichloro-3-(hydroxyimino)-1,3-dihydro-2H-indol-2-one, CALCIUM ION, Calmodulin-2, ... | | Authors: | Nam, Y.W, Zhang, M. | | Deposit date: | 2017-08-07 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A V-to-F substitution in SK2 channels causes Ca2+hypersensitivity and improves locomotion in a C. elegans ALS model.
Sci Rep, 8, 2018
|
|
6KFW
 
 | |
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2L44
 
 | | C-terminal zinc knuckle of the HIVNCp7 | | Descriptor: | C-terminal Zinc Knucle of the HIV-NCP7, ZINC ION | | Authors: | Quintal, S.M.O, Viegas, A.J.M, Cabrita, E.J, Farrell, N.P, Erhardt, S. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Platinated DNA affects zinc finger conformation. Interaction of a platinated single-stranded oligonucleotide and the C-terminal zinc finger of nucleocapsid protein HIVNCp7.
Biochemistry, 51, 2012
|
|
3OA4
 
 | | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125 | | Descriptor: | GLYCEROL, SULFATE ION, ZINC ION, ... | | Authors: | Malashkevich, V.N, Patskovsky, Y, Garrett, S, Foti, R, Toro, R, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-08-04 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | CRYSTAL STRUCTURE OF hypothetical protein BH1468 from Bacillus halodurans C-125
To be Published
|
|
2N1G
 
 | |
2KD7
 
 | | Solution NMR structure of F5/8 type C-terminal domain of a putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium target BtR324B | | Descriptor: | Putative chitobiase | | Authors: | Eletsky, A, Mills, J.L, Lee, H, Lee, D, Jiang, M, Ciccosanti, C, Xiao, R, Nair, R, Everett, J.K, Swapna, G.V.T, Acton, T.B, Rost, B, Montelione, G.T, Prestegard, J.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of F5/8 type C-terminal domain of a putative chitobiase from Bacteroides thetaiotaomicron.
To be Published
|
|
9BH5
 
 | | High-resolution C. elegans 80S ribosome structure - class 1 | | Descriptor: | 18S rRNA, 28S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Sehgal, E, Serrao, V.H.B, Arribere, J. | | Deposit date: | 2024-04-19 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | High-resolution reconstruction of a C. elegans ribosome sheds light on evolutionary dynamics and tissue specificity.
Rna, 30, 2024
|
|
5MTD
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form II | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, TRIETHYLENE GLYCOL, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
2VHD
 
 | | Crystal Structure Of The Di-Haem Cytochrome C Peroxidase From Pseudomonas aeruginosa - Mixed Valence Form | | Descriptor: | CALCIUM ION, CYTOCHROME C551 PEROXIDASE, HEME C | | Authors: | Echalier, A, Brittain, T, Wright, J, Boycheva, S, Mortuza, G.B, Fulop, V, Watmough, N.J. | | Deposit date: | 2007-11-20 | | Release date: | 2008-02-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Redox-Linked Structural Changes Associated with the Formation of a Catalytically Competent Form of the Diheme Cytochrome C Peroxidase from Pseudomonas Aeruginosa
Biochemistry, 47, 2008
|
|
5MTC
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form I | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, ZINC ION | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
1COR
 
 | |
3TXO
 
 | | PKC eta kinase in complex with a naphthyridine | | Descriptor: | 2-methyl-N~1~-[3-(pyridin-4-yl)-2,6-naphthyridin-1-yl]propane-1,2-diamine, Protein kinase C eta type | | Authors: | Stark, W, Rummel, G, Cowan-Jacob, S.W. | | Deposit date: | 2011-09-23 | | Release date: | 2011-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2,6-Naphthyridines as potent and selective inhibitors of the novel protein kinase C isozymes.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1IJS
 
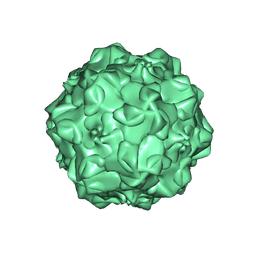 | | CPV (STRAIN D) mutant A300D, complex (VIRAL COAT/DNA), VP2, PH=7.5, T=4 DEGREES C | | Descriptor: | DNA (5'-D(*AP*C)-3'), DNA (5'-D(*CP*CP*AP*CP*CP*CP*CP*AP*A)-3'), PROTEIN (PARVOVIRUS COAT PROTEIN) | | Authors: | Llamas-Saiz, A.L, Agbandje-McKenna, M, Parker, J.S.L, Wahid, A.T.M, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 1996-09-12 | | Release date: | 1996-12-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural analysis of a mutation in canine parvovirus which controls antigenicity and host range.
Virology, 225, 1996
|
|
6M3C
 
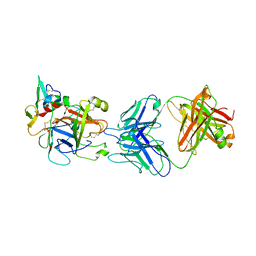 | | hAPC-h1573 Fab complex | | Descriptor: | Vitamin K-dependent protein C heavy chain, Vitamin K-dependent protein C light chain, h1573 Fab H chain, ... | | Authors: | Wang, X, Wang, D, Zhao, X, Egner, U. | | Deposit date: | 2020-03-03 | | Release date: | 2020-07-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Targeted inhibition of activated protein C by a non-active-site inhibitory antibody to treat hemophilia.
Nat Commun, 11, 2020
|
|
5UQE
 
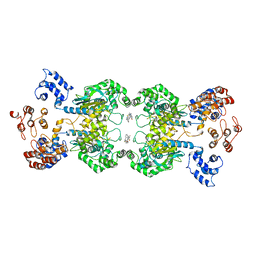 | | Multidomain structure of human kidney-type glutaminase(KGA/GLS) | | Descriptor: | Glutaminase kidney isoform, mitochondrial, N,N'-[sulfanediylbis(ethane-2,1-diyl-1,3,4-thiadiazole-5,2-diyl)]bis(2-phenylacetamide) | | Authors: | Pasquali, C.C, Dias, S.M.G, Ambrosio, A.L.B. | | Deposit date: | 2017-02-08 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The origin and evolution of human glutaminases and their atypical C-terminal ankyrin repeats.
J. Biol. Chem., 292, 2017
|
|
2KXH
 
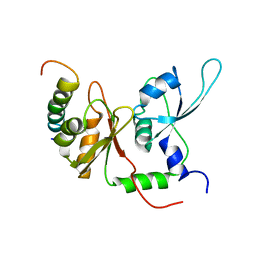 | | Solution structure of the first two RRM domains of FIR in the complex with FBP Nbox peptide | | Descriptor: | Poly(U)-binding-splicing factor PUF60, peptide of Far upstream element-binding protein 1 | | Authors: | Cukier, C.D, Ramos, A, Hollingworth, D, Diaz-Moreno, I, Kelly, G. | | Deposit date: | 2010-05-05 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of FIR-mediated c-myc transcriptional control.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3L4M
 
 | |
