3NP7
 
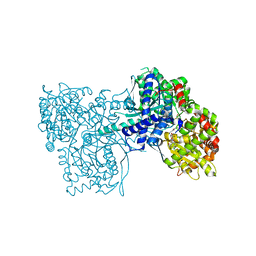 | | Glycogen phosphorylase complexed with 2,5-dihydroxy-3-(beta-D-glucopyranosyl)-chlorobenzene and 2,5-dihydroxy-4-(beta-D-glucopyranosyl)-chlorobenzene | | Descriptor: | (1S)-1,5-anhydro-1-(3-chloro-2,5-dihydroxyphenyl)-D-glucitol, (1S)-1,5-anhydro-1-(4-chloro-2,5-dihydroxyphenyl)-D-glucitol, Glycogen phosphorylase, ... | | Authors: | Alexacou, K.-M. | | Deposit date: | 2010-06-28 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Halogen-substituted (C-beta-D-glucopyranosyl)-hydroquinone regioisomers: synthesis, enzymatic evaluation and their binding to glycogen phosphorylase.
Bioorg.Med.Chem., 19, 2011
|
|
2QRG
 
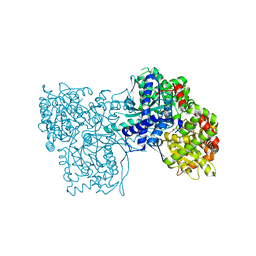 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methoxyphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (5R,7R,8S,9S,10R)-7-(HYDROXYMETHYL)-3-(4-METHOXYPHENYL)-1,6-DIOXA-2-AZASPIRO[4.5]DEC-2-ENE-8,9,10-TRIOL, Glycogen phosphorylase, muscle form | | Authors: | Kizilis, G, Alexacou, K.-M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
2QRQ
 
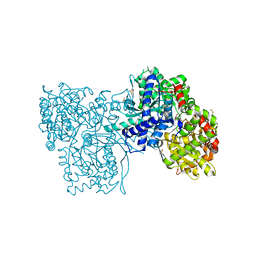 | | Glycogen Phosphorylase b in complex with (1R)-3'-(4-methylphenyl)-spiro[1,5-anhydro-D-glucitol-1,5'-isoxazoline] | | Descriptor: | (3S,5R,7R,8S,9S,10R)-7-(hydroxymethyl)-3-(4-methylphenyl)-1,6-dioxa-2-azaspiro[4.5]decane-8,9,10-triol, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Gizilis, G, Alexacou, K.M, Chrysina, E.D, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-28 | | Release date: | 2008-07-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Glucose-based spiro-isoxazolines: a new family of potent glycogen phosphorylase inhibitors.
Bioorg.Med.Chem., 17, 2009
|
|
3NPA
 
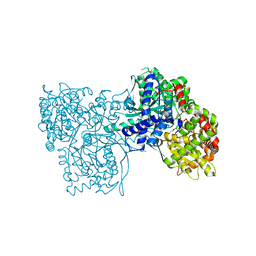 | |
1TL7
 
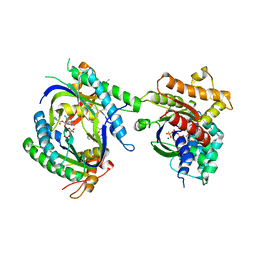 | | Complex Of Gs- With The Catalytic Domains Of Mammalian Adenylyl Cyclase: Complex With 2'(3')-O-(N-methylanthraniloyl)-guanosine 5'-triphosphate and Mn | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | Authors: | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
1SUW
 
 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus in complex with its substrate and product: Insights into the catalysis of NAD kinase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
3OFN
 
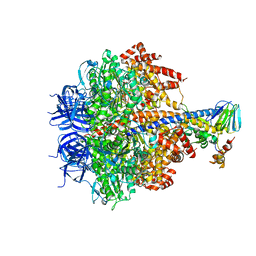 | | Structure of four mutant forms of yeast F1 ATPase: alpha-N67I | | Descriptor: | ATP synthase subunit alpha, ATP synthase subunit beta, ATP synthase subunit delta, ... | | Authors: | Arsenieva, D, Symersky, J, Wang, Y, Pagadala, V, Mueller, D.M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of mutant forms of the yeast f1 ATPase reveal two modes of uncoupling.
J.Biol.Chem., 285, 2010
|
|
1VBG
 
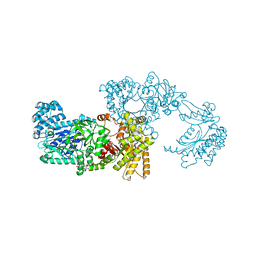 | | Pyruvate Phosphate Dikinase from Maize | | Descriptor: | MAGNESIUM ION, SULFATE ION, pyruvate,orthophosphate dikinase | | Authors: | Nakanishi, T, Nakatsu, T, Matsuoka, M, Sakata, K, Kato, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-26 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of pyruvate phosphate dikinase from maize revealed an alternative conformation in the swiveling-domain motion
Biochemistry, 44, 2005
|
|
1UQ5
 
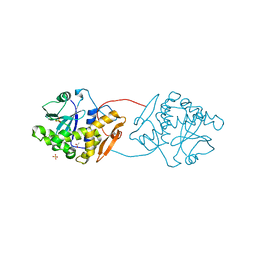 | | RICIN A-CHAIN (RECOMBINANT) N122A MUTANT | | Descriptor: | ACETATE ION, RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
2QN9
 
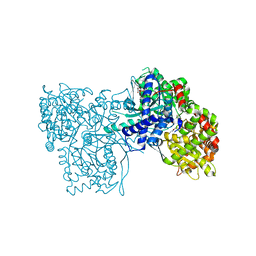 | | Glycogen Phosphorylase in complex with N-4-aminobenzoyl-N'-beta-D-glucopyranosyl urea | | Descriptor: | Glycogen phosphorylase, muscle form, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Tiraidis, C, Alexacou, K.-M, Zographos, S.E, Leonidas, D.D, Oikonomakos, N.G. | | Deposit date: | 2007-07-18 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N-(4-substituted-benzoyl)-N'-(beta-D-glucopyranosyl)ureas, inhibitors of glycogen phosphorylase: synthesis, kinetic and crystallographic evaluation
To be Published
|
|
1UQ4
 
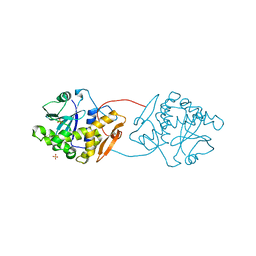 | | RICIN A-CHAIN (RECOMBINANT) R213D MUTANT | | Descriptor: | RICIN, SULFATE ION | | Authors: | Marsden, C.J, Fulop, V. | | Deposit date: | 2003-10-15 | | Release date: | 2004-01-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Effect of Mutations Surrounding and within the Active Site on the Catalytic Activity of Ricin a Chain
Eur.J.Biochem., 271, 2004
|
|
5ON3
 
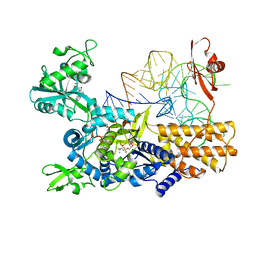 | | Quaternary complex of mutant T252A of E. coli leucyl-tRNA synthetase with tRNA(leu), leucyl-adenylate analogue, and post-transfer editing analogue of leucine in the aminoacylation conformation | | Descriptor: | (2~{S})-~{N}-[(2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-(hydroxymethyl)-4-oxidanyl-oxolan-3-yl]-2-azanyl-4-methyl-pentanamide, 5'-O-(L-leucylsulfamoyl)adenosine, Leucine--tRNA ligase, ... | | Authors: | Palencia, A, Cusack, S. | | Deposit date: | 2017-08-02 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Kinetic Origin of Substrate Specificity in Post-Transfer Editing by Leucyl-tRNA Synthetase.
J. Mol. Biol., 430, 2018
|
|
3OPY
 
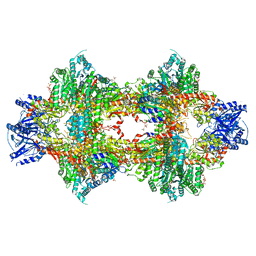 | | Crystal structure of Pichia pastoris phosphofructokinase in the T-state | | Descriptor: | 6-phosphofructo-1-kinase alpha-subunit, 6-phosphofructo-1-kinase beta-subunit, 6-phosphofructo-1-kinase gamma-subunit, ... | | Authors: | Strater, N, Marek, S, Kuettner, E.B, Kloos, M, Keim, A, Bruser, A, Kirchberger, J, Schoneberg, T. | | Deposit date: | 2010-09-02 | | Release date: | 2010-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Molecular architecture and structural basis of allosteric regulation of eukaryotic phosphofructokinases.
Faseb J., 25, 2011
|
|
2XE4
 
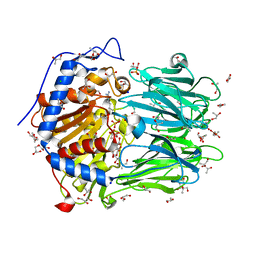 | | Structure of Oligopeptidase B from Leishmania major | | Descriptor: | ANTIPAIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | McLuskey, K, Paterson, N.G, Bland, N.D, Mottram, J.C, Isaacs, N.W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-06 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Leishmania Major Oligopeptidase B Gives Insight Into the Enzymatic Properties of a Trypanosomatid Virulence Factor.
J.Biol.Chem., 285, 2010
|
|
2XOC
 
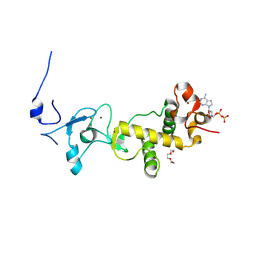 | | C-terminal cysteine-rich domain of human CHFR bound to mADPr | | Descriptor: | ADENINE, ADENOSINE-5'-DIPHOSPHATE, E3 UBIQUITIN-PROTEIN LIGASE CHFR, ... | | Authors: | Oberoi, J, Bayliss, R. | | Deposit date: | 2010-08-11 | | Release date: | 2010-09-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural Basis of Poly(Adp-Ribose) Recognition by the Multizinc Binding Domain of Checkpoint with Forkhead-Associated and Ring Domains (Chfr).
J.Biol.Chem., 285, 2010
|
|
4G0Z
 
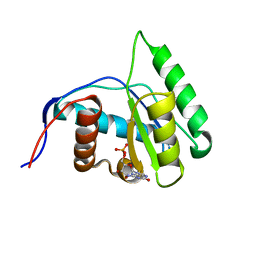 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-10 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
2XQC
 
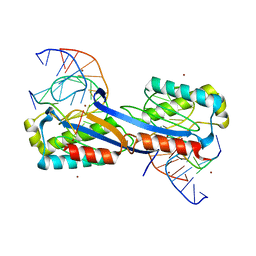 | | DEINOCOCCUS RADIODURANS ISDRA2 TRANSPOSASE COMPLEXED WITH LEFT END RECOGNITION AND CLEAVAGE SITE AND ZN | | Descriptor: | 5'-D(TP*TP*GP*AP*TP*GP)-3', DRA2 TRANSPOSASE LEFT END RECOGNITION SEQUENCE, TRANSPOSASE, ... | | Authors: | Hickman, A.B, James, J.A, Barabas, O, Pasternak, C, Ton-Hoang, B, Chandler, M, Sommer, S, Dyda, F. | | Deposit date: | 2010-09-01 | | Release date: | 2010-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | DNA Recognition and the Precleavage State During Single-Stranded DNA Transposition in D. Radiodurans.
Embo J., 29, 2010
|
|
4G0M
 
 | | Crystal structure of Arabidopsis thaliana AGO2 MID domain | | Descriptor: | Protein argonaute 2, SULFATE ION | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
4G0Q
 
 | | Crystal structure of Arabidopsis thaliana AGO1 MID domain in complex with CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Protein argonaute 1 | | Authors: | Frank, F, Hauver, J, Sonenberg, N, Nagar, B. | | Deposit date: | 2012-07-09 | | Release date: | 2012-07-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs.
Embo J., 31, 2012
|
|
1U0H
 
 | | STRUCTURAL BASIS FOR THE INHIBITION OF MAMMALIAN ADENYLYL CYCLASE BY MANT-GTP | | Descriptor: | 3'-O-(N-METHYLANTHRANILOYL)-GUANOSINE-5'-TRIPHOSPHATE, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Adenylate cyclase, ... | | Authors: | Mou, T.C, Gille, A, Seifert, R.J, Sprang, S.R. | | Deposit date: | 2004-07-13 | | Release date: | 2004-12-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for the inhibition of mammalian membrane adenylyl cyclase by 2 '(3')-O-(N-Methylanthraniloyl)-guanosine 5 '-triphosphate.
J.Biol.Chem., 280, 2005
|
|
1VFL
 
 | | Adenosine deaminase | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2004-04-16 | | Release date: | 2005-08-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Compound Recognition by Adenosine Deaminase
Biochemistry, 44, 2005
|
|
6UK3
 
 | |
6TW0
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12690 (an AOPCP derivative, compound 10 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-azanyl-2-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TWF
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12604 (an AOPCP derivative, compound 21 in publication) in the closed state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, ZINC ION, ... | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-13 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
6TVX
 
 | | Human CD73 (ecto 5'-nucleotidase) in complex with PSB12676 (an AOPCP derivative, compound 9 in paper) in the closed state | | Descriptor: | 5'-nucleotidase, ZINC ION, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,6-bis(azanyl)purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]methylphosphonic acid | | Authors: | Pippel, J, Strater, N. | | Deposit date: | 2020-01-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
