5T7V
 
 | | Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Belousoff, M.J, Lithgow, T, Eyal, Z, Yonath, A, Radjainia, M. | | Deposit date: | 2016-09-06 | | Release date: | 2017-05-31 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in theStaphylococcus aureusRibosome.
MBio, 8, 2017
|
|
5TCU
 
 | | Methicillin sensitive Staphylococcus aureus 70S ribosome | | Descriptor: | 16S RRNA, 23S RRNA, 30S ribosomal protein S10, ... | | Authors: | Eyal, Z, Ahmed, T, Belousoff, N, Mishra, S, Matzov, D, Bashan, A, Zimmerman, E, Lithgow, T, Bhushan, S, Yonath, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-24 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for Linezolid Binding Site Rearrangement in the Staphylococcus aureus Ribosome.
MBio, 8, 2017
|
|
6FJ2
 
 | |
6FJ9
 
 | |
6FJ4
 
 | |
6FJ6
 
 | |
6FID
 
 | | Bovine trypsin solved by S-SAD on ID30B | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | McCarthy, A.A, Mueller-Dieckmann, C. | | Deposit date: | 2018-01-18 | | Release date: | 2018-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ID30B - a versatile beamline for macromolecular crystallography experiments at the ESRF.
J Synchrotron Radiat, 25, 2018
|
|
6FJ8
 
 | |
6GC2
 
 | | AbLIFT: Antibody stability and affinity optimization by computational design of the variable light-heavy chain interface | | Descriptor: | Heavy chain, Light Chain | | Authors: | Warszawski, S, Katz, A, Khmelnitsky, L, Ben Nissan, G, Javitt, G, Dym, O, Unger, T, Knop, O, Diskin, R, Albeck, S, Fass, D, Sharon, M, Fleishman, S.J. | | Deposit date: | 2018-04-17 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Optimizing antibody affinity and stability by the automated design of the variable light-heavy chain interfaces.
Plos Comput.Biol., 15, 2019
|
|
6GBL
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6GBJ
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
6GBK
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
7B38
 
 | | Torpedo californica acetylcholinesterase complexed with Mg+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-29 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7B8E
 
 | | Torpedo californica acetylcholinesterase complexed with Ca+2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(2-ethoxyethoxy)ethoxy]ethanol, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L. | | Deposit date: | 2020-12-12 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
7B2W
 
 | | Torpedo californica acetylcholinesterase complexed with UO2 | | Descriptor: | Acetylcholinesterase, URANYL (VI) ION | | Authors: | Silman, I, Shnyrov, V.L, Ashani, Y, Roth, E, Nicolas, A, Sussman, J.L, Weiner, L. | | Deposit date: | 2020-11-28 | | Release date: | 2021-03-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Torpedo californica acetylcholinesterase is stabilized by binding of a divalent metal ion to a novel and versatile 4D motif.
Protein Sci., 30, 2021
|
|
6GG1
 
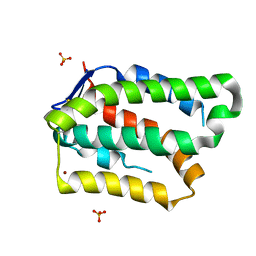 | | Structure of PROSS-edited human interleukin 24 | | Descriptor: | Interleukin-24, NICKEL (II) ION, SULFATE ION | | Authors: | Kolenko, P, Zahradnik, J, Kolarova, L, Schneider, B. | | Deposit date: | 2018-05-02 | | Release date: | 2019-05-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Flexible regions govern promiscuous binding of IL-24 to receptors IL-20R1 and IL-22R1.
Febs J., 286, 2019
|
|
8HDV
 
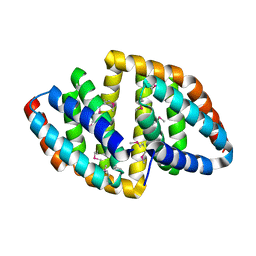 | |
8HDU
 
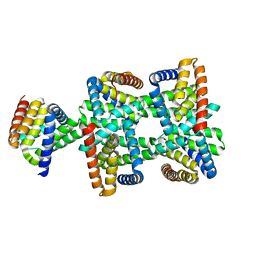 | |
6MKM
 
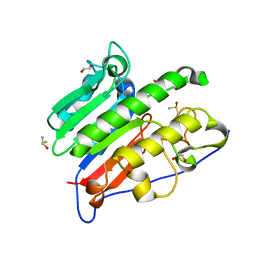 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKO
 
 | |
9DLI
 
 | |
6MKK
 
 | | Crystallographic solvent mapping analysis of DMSO/Mg bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6S0S
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel, ribostamycin B and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, CHLORIDE ION, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Porebski, P.J, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
6S0W
 
 | | The crystal structure of kanamycin B dioxygenase (KanJ) from Streptomyces kanamyceticus in complex with nickel and kanamycin B sulfate | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-DIAMINO-3-[(3-AMINO-3-DEOXY-ALPHA-D-GLUCOPYRANOSYL)OXY]-2-HYDROXYCYCLOHEXYL 2,6-DIAMINO-2,6-DIDEOXY-ALPHA-D-GLUCOPYRANOSIDE, DI(HYDROXYETHYL)ETHER, Kanamycin B dioxygenase, ... | | Authors: | Mrugala, B, Niedzialkowska, E, Minor, W, Borowski, T. | | Deposit date: | 2019-06-18 | | Release date: | 2020-07-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A study on the structure, mechanism, and biochemistry of kanamycin B dioxygenase (KanJ)-an enzyme with a broad range of substrates.
Febs J., 288, 2021
|
|
