7P2M
 
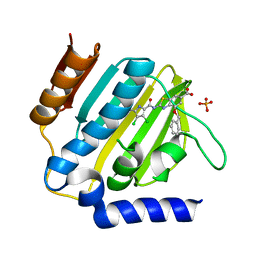 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
5KAF
 
 | | RT XFEL structure of Photosystem II in the dark state at 3.0 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.00001 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
5KAI
 
 | | NH3-bound RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 2.8 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Young, I.D, Ibrahim, M, Chatterjee, R, Gul, S, Koroidov, S, Brewster, A.S, Tran, R, Alonso-Mori, R, Fuller, F, Kroll, T, Michels-Clark, T, Laksmono, H, Sierra, R.G, Stan, C.A, Saracini, C, Bean, M.A, Seuffert, I, Sokaras, D, Weng, T.-C, Hunter, M.S, Aquila, A, Koglin, J.E, Robinson, J, Liang, M, Boutet, S, Lyubimov, A.Y, Uervirojnangkoorn, M, Moriarty, N.W, Liebschner, D, Afonine, P.V, Waterman, D.G, Evans, G, Dobbek, H, Weis, W.I, Brunger, A.T, Zwart, P.H, Adams, P.D, Zouni, A, Messinger, J, Bergmann, U, Sauter, N.K, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2016-06-01 | | Release date: | 2016-11-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.80000925 Å) | | Cite: | Structure of photosystem II and substrate binding at room temperature.
Nature, 540, 2016
|
|
1CLI
 
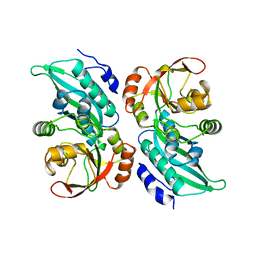 | | X-RAY CRYSTAL STRUCTURE OF AMINOIMIDAZOLE RIBONUCLEOTIDE SYNTHETASE (PURM), FROM THE E. COLI PURINE BIOSYNTHETIC PATHWAY, AT 2.5 A RESOLUTION | | Descriptor: | PROTEIN (PHOSPHORIBOSYL-AMINOIMIDAZOLE SYNTHETASE), SULFATE ION | | Authors: | Li, C, Kappock, T.J, Stubbe, J, Weaver, T.M, Ealick, S.E. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of aminoimidazole ribonucleotide synthetase (PurM), from the Escherichia coli purine biosynthetic pathway at 2.5 A resolution.
Structure Fold.Des., 7, 1999
|
|
6S27
 
 | |
7PTG
 
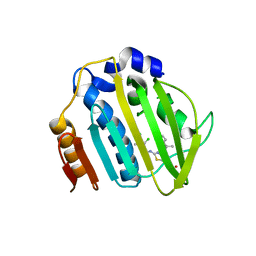 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-27 | | Release date: | 2022-10-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQM
 
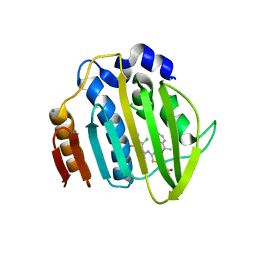 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2888 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
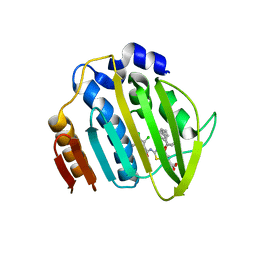 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | Authors: | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
1X92
 
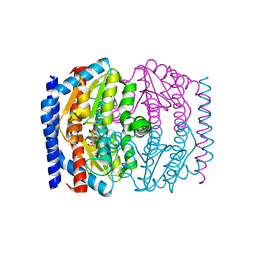 | | CRYSTAL STRUCTURE OF PSEUDOMONAS AERUGINOSA PHOSPHOHEPTOSE ISOMERASE IN COMPLEX WITH REACTION PRODUCT D-GLYCERO-D-MANNOPYRANOSE-7-PHOSPHATE | | Descriptor: | 7-O-phosphono-D-glycero-alpha-D-manno-heptopyranose, PHOSPHOHEPTOSE ISOMERASE | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Joachimiak, A, Edwards, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants.
J.Biol.Chem., 283, 2008
|
|
3WIS
 
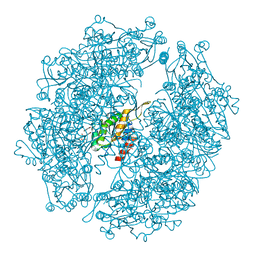 | | Crystal structure of Burkholderia xenovorans DmrB in complex with FMN: A Cubic Protein Cage for Redox Transfer | | Descriptor: | FLAVIN MONONUCLEOTIDE, Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | Authors: | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | Deposit date: | 2013-09-25 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
2I22
 
 | | Crystal structure of Escherichia coli phosphoheptose isomerase in complex with reaction substrate sedoheptulose 7-phosphate | | Descriptor: | D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, Phosphoheptose isomerase | | Authors: | Blakely, K, Zhang, K, DeLeon, G, Wright, G, Junop, M. | | Deposit date: | 2006-08-15 | | Release date: | 2007-08-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
2I2W
 
 | | Crystal Structure of Escherichia Coli Phosphoheptose Isomerase | | Descriptor: | GLYCEROL, Phosphoheptose isomerase | | Authors: | DeLeon, G, Blakely, K, Zhang, K, Wright, G, Junop, M. | | Deposit date: | 2006-08-17 | | Release date: | 2007-08-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants
J.Biol.Chem., 283, 2008
|
|
4MWG
 
 | | Crystal structure of Burkholderia xenovorans DmrB apo form: A Cubic Protein Cage for Redox Transfer | | Descriptor: | Putative dihydromethanopterin reductase (AfpA), SULFATE ION | | Authors: | Bobik, T.A, Cascio, D, Jorda, J, McNamara, D.E, Bustos, C, Wang, T.C, Rasche, M.E, Yeates, T.O. | | Deposit date: | 2013-09-24 | | Release date: | 2014-02-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of dihydromethanopterin reductase, a cubic protein cage for redox transfer
J.Biol.Chem., 289, 2014
|
|
3BJZ
 
 | | Crystal structure of Pseudomonas aeruginosa phosphoheptose isomerase | | Descriptor: | CHLORIDE ION, Phosphoheptose isomerase, SULFATE ION | | Authors: | Walker, J.R, Evdokimova, E, Kudritska, M, Osipiuk, J, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-12-05 | | Release date: | 2007-12-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Function of Sedoheptulose-7-phosphate Isomerase, a Critical Enzyme for Lipopolysaccharide Biosynthesis and a Target for Antibiotic Adjuvants.
J.Biol.Chem., 283, 2008
|
|
1EKH
 
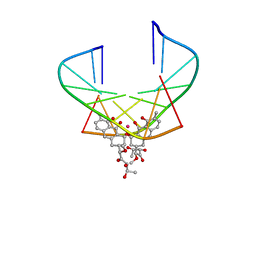 | | NMR STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
1EKI
 
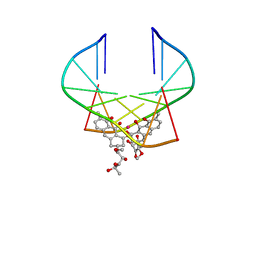 | | AVERAGE SOLUTION STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
1FQT
 
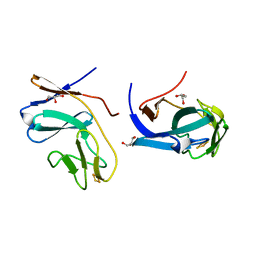 | | CRYSTAL STRUCTURE OF THE RIESKE-TYPE FERREDOXIN ASSOCIATED WITH BIPHENYL DIOXYGENASE | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, RIESKE-TYPE FERREDOXIN OF BIPHENYL DIOXYGENASE | | Authors: | Colbert, C.L, Couture, M.M.-J, Eltis, L.D, Bolin, J.T. | | Deposit date: | 2000-09-06 | | Release date: | 2001-01-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A cluster exposed: structure of the Rieske ferredoxin from biphenyl dioxygenase and the redox properties of Rieske Fe-S proteins.
Structure Fold.Des., 8, 2000
|
|
6Z35
 
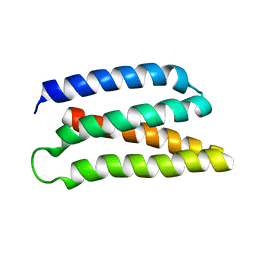 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
6FBA
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
7OGP
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
7OGR
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
6HL7
 
 | | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with mutated active site (R109A/K138A) and N-carbamoyl-L-phosphate bound | | Descriptor: | Aspartate transcarbamoylase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Bosch, S.S, Lunev, S, Wrenger, C, Groves, M.R. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Target Validation of Aspartate Transcarbamoylase fromPlasmodium falciparumby Torin 2.
Acs Infect Dis., 6, 2020
|
|
5J54
 
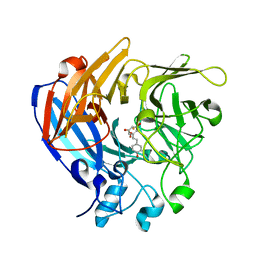 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5J55
 
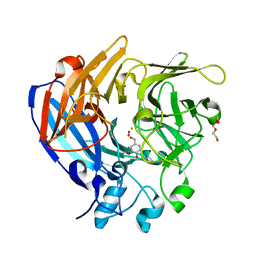 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
5J53
 
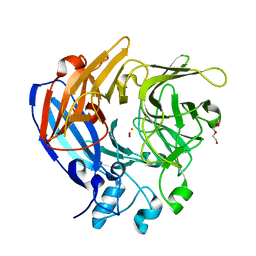 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
