1FOK
 
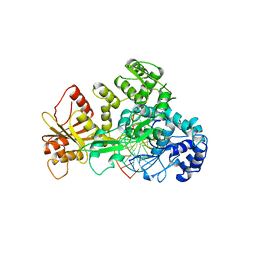 | | STRUCTURE OF RESTRICTION ENDONUCLEASE FOKI BOUND TO DNA | | Descriptor: | DNA (5'-D(*AP*TP*GP*AP*CP*TP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*T P*CP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*GP*AP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*AP*G P*TP*CP*A)-3'), PROTEIN (FOKI RESTRICTION ENDONUCLEASE) | | Authors: | Aggarwal, D.A, Wah, J.A, Hirsch, L.F, Dorner, I, Schildkraut, A.K. | | Deposit date: | 1997-04-18 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the multimodular endonuclease FokI bound to DNA.
Nature, 388, 1997
|
|
6MIH
 
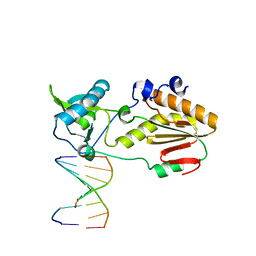 | | Crystal structure of host-guest complex with PC hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*(1WA)P*CP*(DB)P*T)-3'), DNA (5'-D(P*AP*(DS)P*GP*(1W5)P*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIK
 
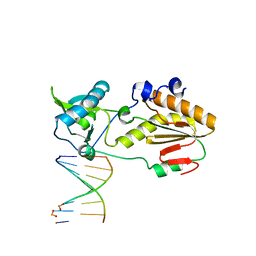 | | Crystal structure of host-guest complex with PP hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), N-terminal fragment of MMLV reverse transcriptase | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
6MIG
 
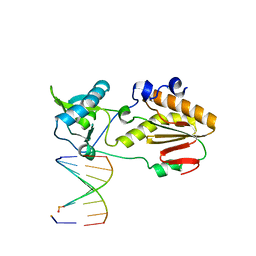 | | Crystal structure of host-guest complex with PB hachimoji DNA | | Descriptor: | DNA (5'-D(*CP*TP*TP*AP*TP*(1WA)P*(1WA)P*(DS))-3'), DNA (5'-D(P*(DB)P*(1W5)P*(1W5)P*AP*TP*AP*AP*G)-3'), Gag-Pol polyprotein | | Authors: | Georgiadis, M.M. | | Deposit date: | 2018-09-19 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Hachimoji DNA and RNA: A genetic system with eight building blocks.
Science, 363, 2019
|
|
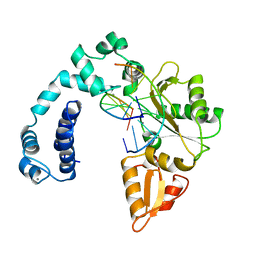
1ZQC
 
 | |
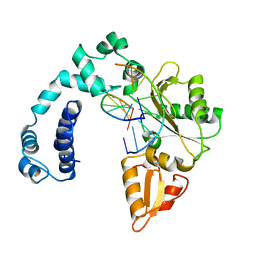
1ZQN
 
 | |
4REC
 
 | | A nuclease-DNA complex form 3 | | Descriptor: | DNA (40-MER), Fanconi-associated nuclease 1, IODIDE ION | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
1NK9
 
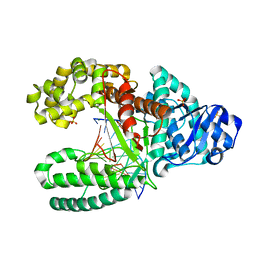 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NKB
 
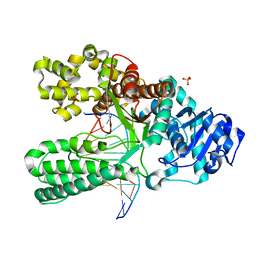 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER THREE ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP, DGTP, AND DTTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1ZQD
 
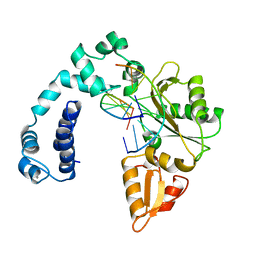 | |
3PY8
 
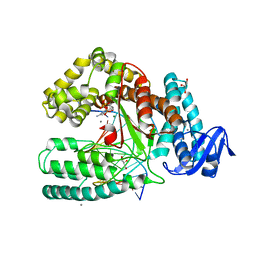 | | Crystal structure of a mutant of the large fragment of DNA polymerase I from thermus aquaticus in a closed ternary complex with DNA and ddCTP | | Descriptor: | 2',3'-DIDEOXYCYTIDINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*A*AP*GP*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Schnur, A, Marx, A, Welte, W, Diederichs, K. | | Deposit date: | 2010-12-12 | | Release date: | 2011-06-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Learning from Directed Evolution: Thermus aquaticus DNA Polymerase Mutants with Translesion Synthesis Activity.
Chembiochem, 12, 2011
|
|
5LCL
 
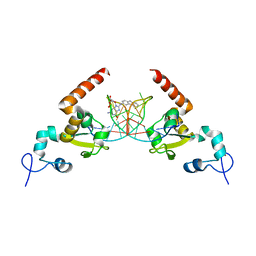 | | STRUCTURE OF the RAD14 DNA-binding domain IN COMPLEX WITH C8-aminofluorene- GUANINE CONTAINING DNA | | Descriptor: | DNA (5'-D(*GP*TP*GP*AP*TP*GP*AP*CP*GP*TP*AP*GP*AP*G)-3'), DNA repair protein RAD14, GCTCTAC(8AF)TCATCA, ... | | Authors: | Schneider, S, Carell, T, Ebert, C, Simon, N. | | Deposit date: | 2016-06-22 | | Release date: | 2017-05-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insights into the Recognition of N(2) -Aryl- and C8-Aryl DNA Lesions by the Repair Protein XPA/Rad14.
Chembiochem, 18, 2017
|
|
1NK8
 
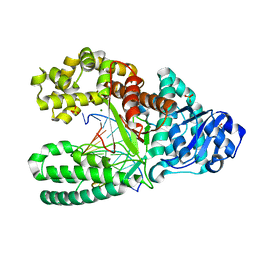 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
4GG4
 
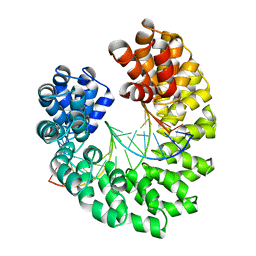 | | Crystal structure of the TAL effector dHax3 bound to specific DNA-RNA hybrid | | Descriptor: | DNA (5'-D(*TP*GP*TP*CP*CP*CP*TP*TP*TP*AP*TP*CP*TP*CP*TP*CP*T)-3'), Hax3, RNA (5'-R(*AP*GP*AP*GP*AP*GP*AP*UP*AP*AP*AP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Yin, P, Deng, D, Yan, C.Y, Pan, X.J, Yan, N, Shi, Y.G. | | Deposit date: | 2012-08-05 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Specific DNA-RNA hybrid recognition by TAL effectors
Cell Rep, 2, 2012
|
|
1HRZ
 
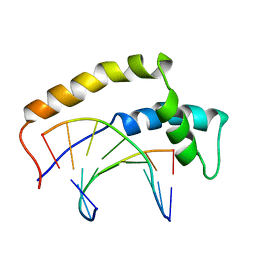 | | THE 3D STRUCTURE OF THE HUMAN SRY-DNA COMPLEX SOLVED BY MULTI-DIMENSIONAL HETERONUCLEAR-EDITED AND-FILTERED NMR | | Descriptor: | DNA (5'-D(*GP*CP*AP*CP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*GP*TP*GP*C)-3'), HUMAN SRY | | Authors: | Clore, G.M, Werner, M.H, Huth, J.R, Gronenborn, A.M. | | Deposit date: | 1995-05-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Cell(Cambridge,Mass.), 81, 1995
|
|
8UVX
 
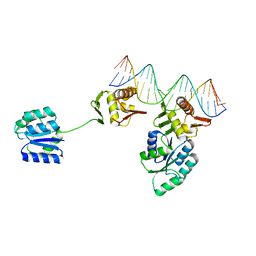 | | CosR DNA bound form I | | Descriptor: | DNA (5'-D(*AP*TP*AP*TP*CP*TP*TP*AP*AP*TP*TP*TP*TP*GP*GP*TP*TP*AP*AP*TP*A)-3'), DNA (5'-D(*TP*AP*TP*TP*AP*AP*CP*CP*AP*AP*AP*AP*TP*TP*AP*AP*GP*AP*TP*AP*T)-3'), DNA-binding response regulator | | Authors: | Zhang, Z. | | Deposit date: | 2023-11-05 | | Release date: | 2024-01-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of DNA recognition of the Campylobacter jejuni CosR regulator.
Mbio, 15, 2024
|
|
6E33
 
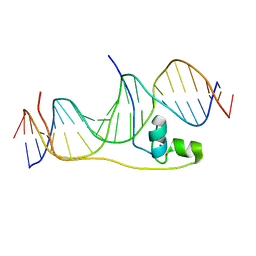 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
4L5N
 
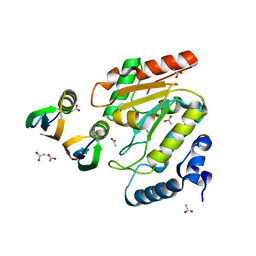 | | Crystallographic Structure of HHV-1 Uracil-DNA Glycosylase complexed with the Bacillus phage PZA inhibitor protein p56 | | Descriptor: | ACETATE ION, Early protein GP1B, Uracil-DNA glycosylase | | Authors: | Cole, A.R, Sapir, O, Ryzhenkova, K, Baltulionis, G, Hornyak, P, Savva, R. | | Deposit date: | 2013-06-11 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Architecturally diverse proteins converge on an analogous mechanism to inactivate Uracil-DNA glycosylase.
Nucleic Acids Res., 41, 2013
|
|
7F4Y
 
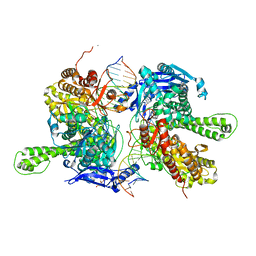 | | Crystal structure of replisomal dimer of DNA polymerase from bacteriophage RB69 with DNA duplexes | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*AP*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*C)-3'), ... | | Authors: | Youn, H.-S, Park, J, An, J.Y, Lee, Y, Eom, S.H, Wang, J. | | Deposit date: | 2021-06-21 | | Release date: | 2021-07-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of New Binary and Ternary DNA Polymerase Complexes From Bacteriophage RB69.
Front Mol Biosci, 8, 2021
|
|
4JWM
 
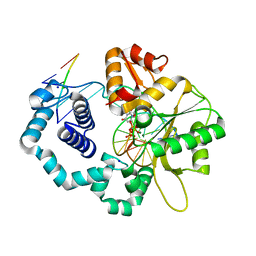 | | Ternary complex of D256E mutant of DNA Polymerase Beta | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, DNA (5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), ... | | Authors: | Batra, V.K, Perera, L, Ping, L, Shock, D.D, Beard, W.A, Pedersen, L.C, Pedersen, L.G, Wilson, S.H. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Amino Acid Substitution in the Active Site of DNA Polymerase beta Explains the Energy Barrier of the Nucleotidyl Transfer Reaction.
J.Am.Chem.Soc., 135, 2013
|
|
2HWV
 
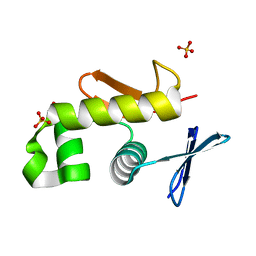 | | Crystal structure of an essential response regulator DNA binding domain, VicRc in Enterococcus faecalis, a member of the YycF subfamily. | | Descriptor: | DNA-binding response regulator VicR, SULFATE ION | | Authors: | Trinh, C.H, Liu, Y, Phillips, S.E.V, Phillips-Jones, M.K. | | Deposit date: | 2006-08-02 | | Release date: | 2007-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the response regulator VicR DNA-binding domain.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4PPX
 
 | | DNA Polymerase Beta E295K with Spiroiminodihydantoin in Templating Position | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*(SDH)P*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(*GP*TP*CP*GP*G)-3', ... | | Authors: | Eckenroth, B.E, Doublie, S. | | Deposit date: | 2014-02-27 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structure of DNA Polymerase beta with DNA Containing the Base Lesion Spiroiminodihydantoin in a Templating Position.
Biochemistry, 53, 2014
|
|
8K86
 
 | | Crystal structure of NFIL3 in complex with TTATGTAA DNA | | Descriptor: | DNA (5'-D(*CP*AP*TP*TP*AP*TP*GP*TP*AP*AP*CP*G)-3'), DNA (5'-D(*CP*GP*TP*TP*AP*CP*AP*TP*AP*AP*TP*G)-3'), Nuclear factor interleukin-3-regulated protein | | Authors: | Min, J.R, Chen, S.Z, Liu, K. | | Deposit date: | 2023-07-28 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for specific DNA sequence recognition by the transcription factor NFIL3.
J.Biol.Chem., 300, 2024
|
|
1MDY
 
 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
5IIO
 
 | | Crystal structure of the DNA polymerase lambda binary complex | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*AP*C)-3'), DNA (5'-D(*CP*GP*GP*CP*(8OG)P*GP*TP*AP*CP*TP*G)-3'), DNA (5'-D(P*GP*CP*CP*G)-3'), ... | | Authors: | Burak, M.J, Guja, K.E, Garcia-Diaz, M. | | Deposit date: | 2016-03-01 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A fidelity mechanism in DNA polymerase lambda promotes error-free bypass of 8-oxo-dG.
Embo J., 35, 2016
|
|
