5WXY
 
 | | Crystal structure of Microcystis aeruginosa PCC 7806 aspartate racemase in complex with L-aspartate | | Descriptor: | ASPARTIC ACID, McyF | | Authors: | Cao, D.D, Zhou, K, Jiang, Y.L, Zhou, C.Z. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Structural insights into the catalysis and substrate specificity of cyanobacterial aspartate racemase McyF.
Biochem.Biophys.Res.Commun., 514, 2019
|
|
2J4W
 
 | | Structure of a Plasmodium vivax apical membrane antigen 1-Fab F8.12.19 complex | | Descriptor: | APICAL MEMBRANE ANTIGEN 1, FAB FRAGMENT OF MONOCLONAL ANTIBODY F8.12.19 | | Authors: | Igonet, S, Vulliez-Le Normand, B, Faure, G, Riottot, M.M, Kocken, C.H.M, Thomas, A.W, Bentley, G.A. | | Deposit date: | 2006-09-07 | | Release date: | 2007-01-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cross-Reactivity Studies of an Anti-Plasmodium Vivax Apical Membrane Antigen 1 Monoclonal Antibody: Binding and Structural Characterisation.
J.Mol.Biol., 366, 2007
|
|
2VKL
 
 | | X-ray crystal structure of the intracellular Chorismate mutase from Mycobactrerium Tuberculosis in complex with malate | | Descriptor: | D-MALATE, RV0948C/MT0975 | | Authors: | Okvist, M, Roderer, K, Sasso, S, Kast, P, Krengel, U. | | Deposit date: | 2007-12-20 | | Release date: | 2008-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and Function of a Complex between Chorismate Mutase and Dahp Synthase: Efficiency Boost for the Junior Partner.
Embo J., 28, 2009
|
|
2JFV
 
 | |
6FAH
 
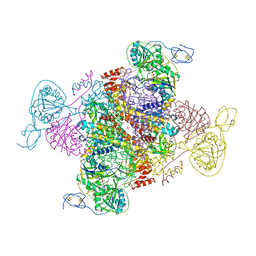 | | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction | | Descriptor: | Caffeyl-CoA reductase-Etf complex subunit CarC, Caffeyl-CoA reductase-Etf complex subunit CarD, Caffeyl-CoA reductase-Etf complex subunit CarE, ... | | Authors: | Demmer, J.K, Bertsch, J, Oeppinger, C, Wohlers, H, Kayastha, K, Demmer, U, Ermler, U, Mueller, V. | | Deposit date: | 2017-12-15 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.133 Å) | | Cite: | Molecular basis of the flavin-based electron-bifurcating caffeyl-CoA reductase reaction.
FEBS Lett., 592, 2018
|
|
2JFP
 
 | |
2JFY
 
 | |
2I57
 
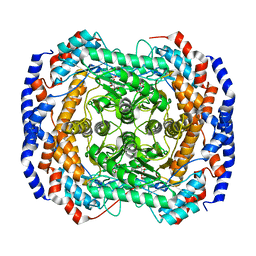 | | Crystal Structure of L-Rhamnose Isomerase from Pseudomonas stutzeri in Complex with D-Allose | | Descriptor: | D-ALLOSE, L-rhamnose isomerase, ZINC ION | | Authors: | Yoshida, H, Yamada, M, Takada, G, Izumori, K, Kamitori, S. | | Deposit date: | 2006-08-24 | | Release date: | 2006-12-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
J.Mol.Biol., 365, 2007
|
|
5A1R
 
 | |
2IHN
 
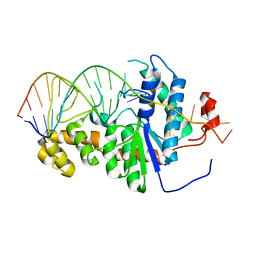 | | Co-crystal of Bacteriophage T4 RNase H with a fork DNA substrate | | Descriptor: | 5'-D(*CP*TP*AP*AP*CP*TP*TP*TP*GP*AP*GP*GP*CP*AP*GP*AP*CP*C)-3', 5'-D(*GP*GP*TP*CP*TP*GP*CP*CP*TP*CP*AP*AP*GP*AP*CP*GP*GP*TP*AP*GP*TP*CP*AP*A)-3', Ribonuclease H | | Authors: | Devos, J.M, Mueser, T.C. | | Deposit date: | 2006-09-26 | | Release date: | 2007-08-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of bacteriophage T4 5' nuclease in complex with a branched DNA reveals how FEN-1 family nucleases bind their substrates.
J.Biol.Chem., 282, 2007
|
|
7E3U
 
 | | Crystal structure of the Pseudomonas aeruginosa dihydropyrimidinase complexed with 5-AU | | Descriptor: | 5-AMINO-1H-PYRIMIDINE-2,4-DIONE, D-hydantoinase/dihydropyrimidinase, ZINC ION | | Authors: | Yang, Y.C, Luo, R.H, Huang, Y.H, Huang, C.Y, Lin, E.S. | | Deposit date: | 2021-02-09 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Molecular Insights into How the Dimetal Center in Dihydropyrimidinase Can Bind the Thymine Antagonist 5-Aminouracil: A Different Binding Mode from the Anticancer Drug 5-Fluorouracil.
Bioinorg Chem Appl, 2022, 2022
|
|
2JFW
 
 | |
4B0N
 
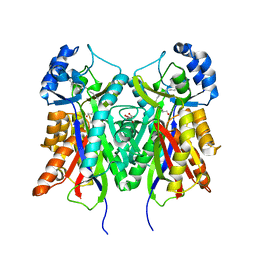 | | Crystal structure of PKS-I from the brown algae Ectocarpus siliculosus | | Descriptor: | ARACHIDONIC ACID, MALONIC ACID, POLYKETIDE SYNTHASE III | | Authors: | Leroux, C, Meslet-Cladiere, L, Delage, L, Goulitquer, S, Leblanc, C, Ar Gall, E, Stiger-Pouvreau, V, Potin, P, Czjzek, M. | | Deposit date: | 2012-07-03 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure/Function Analysis of a Type III Polyketide Synthase in the Brown Alga Ectocarpus Siliculosus Reveals a Biochemical Pathway in Phlorotannin Monomer Biosynthesis.
Plant Cell, 25, 2013
|
|
2JFZ
 
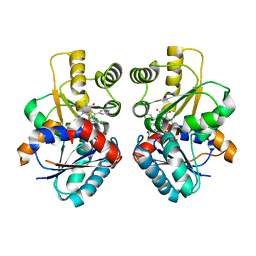 | |
2JFQ
 
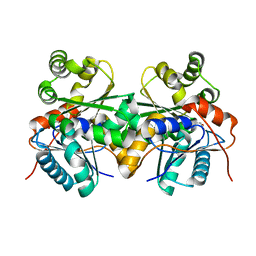 | |
2NNI
 
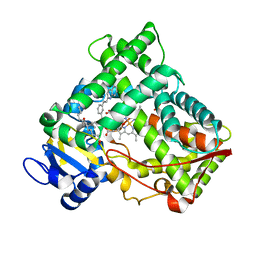 | | CYP2C8dH complexed with montelukast | | Descriptor: | Cytochrome P450 2C8, MONTELUKAST, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
5X1A
 
 | |
2JJN
 
 | | Structure of closed cytochrome P450 EryK | | Descriptor: | CYTOCHROME P450 113A1, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JJO
 
 | | Structure of cytochrome P450 EryK in complex with its natural substrate erD | | Descriptor: | CYTOCHROME P450 113A1, Erythromycin D, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Savino, C, Sciara, G, Miele, A.E, Kendrew, S.G, Vallone, B. | | Deposit date: | 2008-04-15 | | Release date: | 2009-07-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Investigating the Structural Plasticity of a Cytochrome P450: Three-Dimensional Structures of P450 Eryk and Binding to its Physiological Substrate.
J.Biol.Chem., 284, 2009
|
|
2JFU
 
 | |
2I3C
 
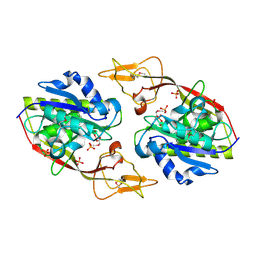 | | Crystal Structure of an Aspartoacylase from Homo Sapiens | | Descriptor: | Aspartoacylase, PHOSPHATE ION, ZINC ION | | Authors: | Bitto, E, Wesenberg, G.E, Phillips Jr, G.N, Mccoy, J.G, Bingman, C.A, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-08-17 | | Release date: | 2006-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of aspartoacylase, the brain enzyme impaired in Canavan disease.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4CHE
 
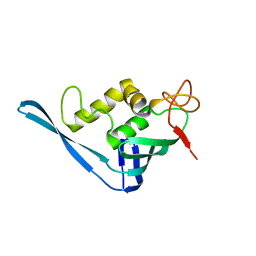 | | Crystal structure of the putative cap-binding domain of the PB2 subunit of Thogoto virus polymerase | | Descriptor: | POLYMERASE BASIC PROTEIN 2 | | Authors: | Guilligay, D, Kadlec, J, Crepin, T, Lunardi, T, Bouvier, D, Kochs, G, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 2013-12-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparative Structural and Functional Analysis of Orthomyxovirus Polymerase CAP-Snatching Domains.
Plos One, 9, 2014
|
|
2NNH
 
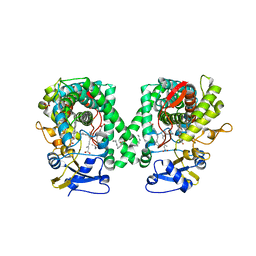 | | CYP2C8dH complexed with 2 molecules of 9-cis retinoic acid | | Descriptor: | (9cis)-retinoic acid, Cytochrome P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Stout, C.D, Johnson, E.F. | | Deposit date: | 2006-10-24 | | Release date: | 2007-10-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J.Biol.Chem., 283, 2008
|
|
4Z3D
 
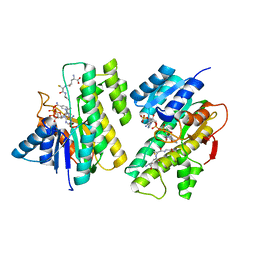 | |
5YM3
 
 | | CYP76AH1-4pi from salvia miltiorrhiza | | Descriptor: | 4-PHENYL-1H-IMIDAZOLE, Ferruginol synthase, MANGANESE (II) ION, ... | | Authors: | Chang, Z. | | Deposit date: | 2017-10-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Crystal structure of CYP76AH1 in 4-PI-bound state from Salvia miltiorrhiza.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
