5EH5
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5EHV
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | (~{E})-3-[3-[[3-(2-hydroxy-2-oxoethyl)phenyl]methoxy]phenyl]prop-2-enoic acid, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.208 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
8HAD
 
 | |
8HAC
 
 | |
1HNI
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN A COMPLEX WITH THE NONNUCLEOSIDE INHIBITOR ALPHA-APA R 95845 AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | (2-ACETYL-5-METHYLANILINO)(2,6-DIBROMOPHENYL)ACETAMIDE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Ding, J, Das, K, Arnold, E. | | Deposit date: | 1995-02-28 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of HIV-1 reverse transcriptase in a complex with the non-nucleoside inhibitor alpha-APA R 95845 at 2.8 A resolution.
Structure, 3, 1995
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
6Z9V
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting IIGWMWIPV | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9X
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLS (t-butyl)Y FGTPT | | Descriptor: | Beta-2-microglobulin, LEU-LEU-SER-TUR-PHE-GLY-THR-PRO-THR, MHC class I antigen, ... | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z9W
 
 | | Human Class I Major Histocompatibility Complex, A02 allele, presenting LLGWVFAQV | | Descriptor: | Beta-2-microglobulin, LEU-LEU-GLY-TRP-VAL-PHE-ALA-GLN-VAL, MHC class I antigen | | Authors: | Rizkallah, P.J, Man, S, Redman, J.E. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthetic Peptides with Inadvertent Chemical Modifications Can Activate Potentially Autoreactive T Cells.
J Immunol., 207, 2021
|
|
6Z1K
 
 | |
6Z1L
 
 | |
5EH8
 
 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | (~{E})-3-(4-methoxyphenyl)but-2-enoic acid, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.381 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
3SGA
 
 | |
6SNT
 
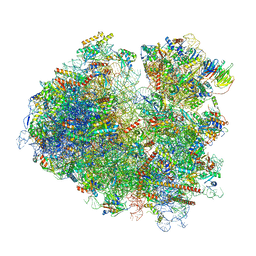 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
3CYR
 
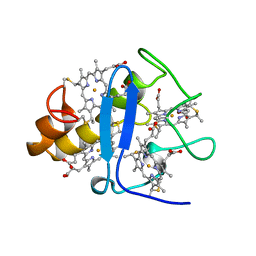 | | CYTOCHROME C3 FROM DESULFOVIBRIO DESULFURICANS ATCC 27774P | | Descriptor: | CYTOCHROME C3, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Simoes, P, Matias, P.M, Morais, J, Wilson, K, Dauter, Z, Carrondo, M.A. | | Deposit date: | 1997-07-24 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refinement of the Three-Dimensional Structures of Cytochrome C3 from Desulfovibrio Vulgaris Hildenborough at 1.67 Angstroms Resolution and from Desulfovibrio Desulfuricans Atcc 27774 at 1.6 Angstroms Resolution
Inorg.Chim.Acta., 273, 1998
|
|
3SQC
 
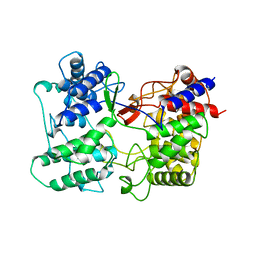 | | SQUALENE-HOPENE CYCLASE | | Descriptor: | SQUALENE--HOPENE CYCLASE | | Authors: | Wendt, K.U, Schulz, G.E. | | Deposit date: | 1998-09-04 | | Release date: | 1999-05-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of the membrane protein squalene-hopene cyclase at 2.0 A resolution.
J.Mol.Biol., 286, 1999
|
|
8IF2
 
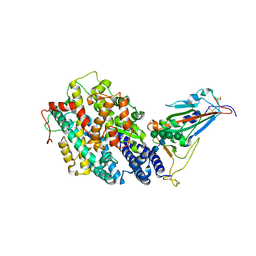 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Omicron BQ.1.1 variant spike protein in complex with its receptor ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Kimura, K, Suzuki, T, Hashiguchi, T. | | Deposit date: | 2023-02-17 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Convergent evolution of SARS-CoV-2 Omicron subvariants leading to the emergence of BQ.1.1 variant.
Nat Commun, 14, 2023
|
|
7WTQ
 
 | | Cryo-EM structure of a yeast pre-40S ribosomal subunit - State Tsr1-2 (without Rps2) | | Descriptor: | 18S rRNA, 18S rRNA (guanine(1575)-N(7))-methyltransferase, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Thoms, M, Ameismeier, M, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-19 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The nucleoplasmic phase of pre-40S formation prior to nuclear export.
Nucleic Acids Res., 50, 2022
|
|
6ZPK
 
 | | Crystal structure of the unconventional kinetochore protein Trypanosoma brucei KKT4 BRCT domain | | Descriptor: | GLYCEROL, SULFATE ION, Trypanosoma brucei KKT4 463-645 | | Authors: | Ludzia, P, Lowe, E.D, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
6ZPO
 
 | | bovine ATP synthase monomer state 1 (combined) | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Spikes, T.E, Montgomery, M.G, Walker, J.E. | | Deposit date: | 2020-07-09 | | Release date: | 2020-09-09 | | Last modified: | 2020-09-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of the dimeric ATP synthase from bovine mitochondria.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZZZ
 
 | |
7T02
 
 | |
7A4N
 
 | | Cryo-EM structure of a prefusion stabilized SARS-CoV-2 Spike (D614N, R682S, R685G, A892P, A942P and V987P)(S-closed trimer) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Rutten, L, Renault, L.L.R, Juraszek, J, Langedijk, J.P.M. | | Deposit date: | 2020-08-20 | | Release date: | 2020-11-04 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Stabilizing the closed SARS-CoV-2 spike trimer.
Nat Commun, 12, 2021
|
|
6ZNS
 
 | | Crystal Structure of DUF1998 helicase MrfA | | Descriptor: | Uncharacterized ATP-dependent helicase YprA, ZINC ION | | Authors: | Roske, J.J, Liu, S, Loll, B, Neu, U, Wahl, M.C. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | A skipping rope translocation mechanism in a widespread family of DNA repair helicases.
Nucleic Acids Res., 49, 2021
|
|
1P4S
 
 | |
