3E9I
 
 | | Lysyl-tRNA synthetase from Bacillus stearothermophilus complexed with L-Lysine hydroxamate-AMP | | Descriptor: | 5'-O-{(R)-hydroxy[(L-lysylamino)oxy]phosphoryl}adenosine, Lysyl-tRNA synthetase, MAGNESIUM ION, ... | | Authors: | Sakurama, H, Takita, T, Mikami, B, Itoh, T, Yasukawa, K, Inouye, K. | | Deposit date: | 2008-08-22 | | Release date: | 2009-07-14 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Two crystal structures of lysyl-tRNA synthetase from Bacillus stearothermophilus in complex with lysyladenylate-like compounds: insights into the irreversible formation of the enzyme-bound adenylate of L-lysine hydroxamate
J.Biochem., 145, 2009
|
|
3RDR
 
 | | Structure of the catalytic domain of XlyA | | Descriptor: | CHLORIDE ION, N-acetylmuramoyl-L-alanine amidase XlyA, ZINC ION | | Authors: | Low, L.Y, Liddington, R.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Role of net charge on catalytic domain and influence of cell wall binding domain on bactericidal activity, specificity, and host range of phage lysins.
J.Biol.Chem., 286, 2011
|
|
3DMJ
 
 | | CRYSTAL STRUCTURE OF HIV-1 V106A and Y181C MUTANT REVERSE TRANSCRIPTASE IN COMPLEX WITH GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-07-01 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
3DPL
 
 | |
2XVU
 
 | |
3RDH
 
 | | X-ray induced covalent inhibition of 14-3-3 | | Descriptor: | 14-3-3 protein zeta/delta, 4-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]benzoic acid, NICKEL (II) ION | | Authors: | Horton, J.R, Upadhyay, A.K, Fu, H, Cheng, X. | | Deposit date: | 2011-04-01 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Discovery and structural characterization of a small molecule 14-3-3 protein-protein interaction inhibitor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3DTO
 
 | | Crystal structure of the metal-dependent HD domain-containing hydrolase BH2835 from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR130. | | Descriptor: | BH2835 protein | | Authors: | Forouhar, F, Su, M, Seetharaman, J, Janjua, H, Fang, Y, Xiao, R, Cunningham, K, Ma, L.-C, Owen, L.A, Wang, D, Tong, S, Everett, J.K, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the metal-dependent HD domain-containing hydrolase BH2835 from Bacillus halodurans, Northeast Structural Genomics Consortium Target BhR130.
To be Published
|
|
2XPJ
 
 | |
3DYU
 
 | |
3RP2
 
 | | THE STRUCTURE OF RAT MAST CELL PROTEASE II AT 1.9-ANGSTROMS RESOLUTION | | Descriptor: | RAT MAST CELL PROTEASE II | | Authors: | Reynolds, R, Remington, S, Weaver, L, Fischer, R, Anderson, W, Ammon, H, Matthews, B. | | Deposit date: | 1984-09-10 | | Release date: | 1984-10-29 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of rat mast cell protease II at 1.9-A resolution.
Biochemistry, 27, 1988
|
|
3DMY
 
 | |
2XWU
 
 | | CRYSTAL STRUCTURE OF IMPORTIN 13 - UBC9 COMPLEX | | Descriptor: | IMPORTIN13, SUMO-CONJUGATING ENZYME UBC9 | | Authors: | Gruenwald, M, Bono, F. | | Deposit date: | 2010-11-05 | | Release date: | 2010-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Importin13-Ubc9 Complex: Nuclear Import and Release of a Key Regulator of Sumoylation.
Embo J., 30, 2011
|
|
3DNM
 
 | |
3DM2
 
 | | Crystal structure of HIV-1 K103N mutant reverse transcriptase in complex with GW564511. | | Descriptor: | N-{4-[amino(dihydroxy)-lambda~4~-sulfanyl]-2-methylphenyl}-2-(4-chloro-2-{[3-fluoro-5-(trifluoromethyl)phenyl]carbonyl}phenoxy)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Chamberlain, P.P, Stammers, D.K. | | Deposit date: | 2008-06-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
4K1J
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
3DVV
 
 | |
3RG9
 
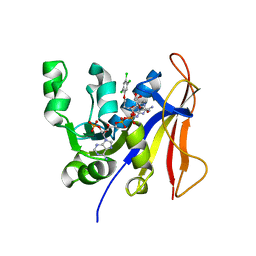 | | Trypanosoma brucei dihydrofolate reductase (TbDHFR) in complex with WR99210 | | Descriptor: | 6,6-DIMETHYL-1-[3-(2,4,5-TRICHLOROPHENOXY)PROPOXY]-1,6-DIHYDRO-1,3,5-TRIAZINE-2,4-DIAMINE, Bifunctional dihydrofolate reductase-thymidylate synthase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Vanichtanankul, J, Yuvaniyama, J, Yuthavong, Y. | | Deposit date: | 2011-04-08 | | Release date: | 2011-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trypanosomal dihydrofolate reductase reveals natural antifolate resistance
Acs Chem.Biol., 6, 2011
|
|
3REV
 
 | | Crystal structure of human alloreactive tcr nb20 | | Descriptor: | 1,2-ETHANEDIOL, TCR NB20 ALPHA CHAIN, TCR NB20 BETA CHAIN | | Authors: | Wood, A, Mohammed, F, Salim, M, Tranter, A, Rickinson, A.B, Moss, P.A.H, Stauss, H.J, Steven, N.M, Willcox, B.E. | | Deposit date: | 2011-04-05 | | Release date: | 2012-01-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and energetic evidence for highly peptide-specific tumor antigen targeting via allo-MHC restriction.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2W35
 
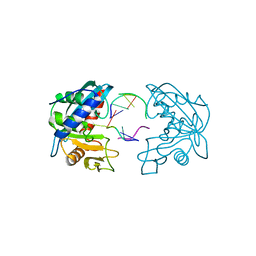 | | Structures of endonuclease V with DNA reveal initiation of deaminated adenine repair | | Descriptor: | 5'-D(*AP*GP*CP*CP*GP*TP)-3', 5'-D(*AP*TP*GP*CP*GP*AP*CP*IP*GP)-3', Endonuclease V, ... | | Authors: | Dalhus, B, Arvai, A.S, Rosnes, I, Olsen, O.E, Backe, P.H, Alseth, I, Gao, H, Cao, W, Tainer, J.A, Bjoras, M. | | Deposit date: | 2008-11-06 | | Release date: | 2009-01-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structures of Endonuclease V with DNA Reveal Initiation of Deaminated Adenine Repair.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2W26
 
 | | Factor Xa in complex with BAY59-7939 | | Descriptor: | 5-chloro-N-({(5S)-2-oxo-3-[4-(3-oxomorpholin-4-yl)phenyl]-1,3-oxazolidin-5-yl}methyl)thiophene-2-carboxamide, ACTIVATED FACTOR XA HEAVY CHAIN, CALCIUM ION | | Authors: | Roehrig, S, Straub, A, Pohlmann, J, Lampe, T, Pernerstorfer, J, Schlemmer, K, Reinemer, P, Perzborn, E, Schaefer, M. | | Deposit date: | 2008-10-24 | | Release date: | 2008-11-11 | | Last modified: | 2021-04-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of the Novel Antithrombotic Agent 5-Chloro-N-({(5S)-2-Oxo-3- [4-(3-Oxomorpholin-4-Yl)Phenyl]-1,3-Oxazolidin-5-Yl}Methyl)Thiophene-2- Carboxamide (Bay 59-7939): An Oral, Direct Factor Xa Inhibitor.
J.Med.Chem., 48, 2005
|
|
2W2D
 
 | | Crystal Structure of a Catalytically Active, Non-toxic Endopeptidase Derivative of Clostridium botulinum Toxin A | | Descriptor: | ACETATE ION, BOTULINUM NEUROTOXIN A HEAVY CHAIN, BOTULINUM NEUROTOXIN A LIGHT CHAIN, ... | | Authors: | Masuyer, G, Thiyagarajan, N, James, P.L, Marks, P.M.H, Chaddock, J.A, Acharya, K.R. | | Deposit date: | 2008-10-29 | | Release date: | 2009-03-24 | | Last modified: | 2022-05-04 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal Structure of a Catalytically Active, Non-Toxic Endopeptidase Derivative of Clostridium Botulinum Toxin A.
Biochem.Biophys.Res.Commun., 381, 2009
|
|
2VYU
 
 | | CRYSTAL STRUCTURE OF CHOLINE BINDING PROTEIN F FROM STREPTOCOCCUS PNEUMONIAE IN THE PRESENCE OF A PEPTIDOGLYCAN ANALOGUE (TETRASACCHARIDE-PENTAPEPTIDE) | | Descriptor: | CHOLINE BINDING PROTEIN F, CHOLINE ION | | Authors: | Perez-Dorado, I, Molina, R, Hermoso, J.A, Mobashery, S. | | Deposit date: | 2008-07-28 | | Release date: | 2009-02-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structure of Cbpf, a Bifunctional Choline-Binding Protein and Autolysis Regulator from Streptococcus Pneumoniae.
Embo Rep., 10, 2009
|
|
3DYF
 
 | | T. Brucei Farnesyl Diphosphate Synthase Complexed with Bisphosphonate BPH-461 and Isopentyl Diphosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 3-FLUORO-1-(2-HYDROXY-2,2-DIPHOSPHONOETHYL)PYRIDINIUM, ACETATE ION, ... | | Authors: | Cao, R, Gao, Y, Robinson, H, Goddard, A, Oldfield, E. | | Deposit date: | 2008-07-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Lipophilic bisphosphonates as dual farnesyl/geranylgeranyl diphosphate synthase inhibitors: an X-ray and NMR investigation.
J.Am.Chem.Soc., 131, 2009
|
|
2VP3
 
 | |
2VS2
 
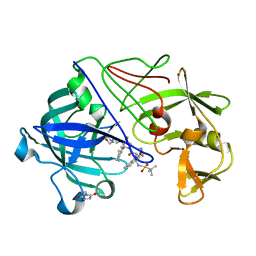 | | Neutron diffraction structure of endothiapepsin in complex with a gem- diol inhibitor. | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Coates, L, Tuan, H.-F, Tomanicek, S, Kovalevsky, A, Mustyakimov, M, Erskine, P, Cooper, J. | | Deposit date: | 2008-04-17 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-15 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | The Catalytic Mechanism of an Aspartic Proteinase Explored with Neutron and X-Ray Diffraction
J.Am.Chem.Soc., 130, 2008
|
|
