3QBW
 
 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to adenosine diphosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
3OFW
 
 | | Crystal structure of recombinant Kunitz Type serine protease Inhibitor-1 from the Carribean sea anemone stichodactyla helianthus | | Descriptor: | CHLORIDE ION, Kunitz-type proteinase inhibitor SHPI-1 | | Authors: | Garcia-Fernandez, R, Redecke, L, Pons, T, Perbandt, M, Talavera, A, Gil, D, Gonzalez, Y, de los Angeles Chavez, M, Betzel, C. | | Deposit date: | 2010-08-16 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the recombinant BPTI/Kunitz-type inhibitor rShPI-1A from the marine invertebrate Stichodactyla helianthus.
Acta Crystallogr.,Sect.F, 68, 2012
|
|
3OGN
 
 | |
4ISW
 
 | | Crystal Structure of Phosphorylated C.elegans Thymidylate Synthase in Complex with dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Wilk, P, Dowiercial, A, Banaszak, K, Jarmula, A, Rypniewski, W, Rode, W. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-11 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Crystal structure of phosphoramide-phosphorylated thymidylate synthase reveals pSer127, reflecting probably pHis to pSer phosphotransfer.
Bioorg.Chem., 52C, 2013
|
|
3QBX
 
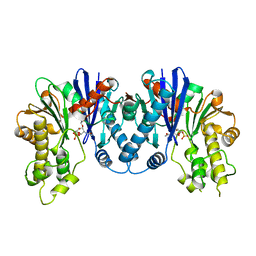 | | Crystal structure of pseudomonas aeruginosa 1,6-anhydro-n-actetylmuramic acid kinase (ANMK) bound to 1,6-anhydro-n-actetylmuramic acid | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, SULFATE ION | | Authors: | Bacik, J.P, Martin, D.R, Mark, B.L. | | Deposit date: | 2011-01-14 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular Basis of 1,6-Anhydro Bond Cleavage and Phosphoryl Transfer by Pseudomonas aeruginosa 1,6-Anhydro-N-acetylmuramic Acid Kinase.
J.Biol.Chem., 286, 2011
|
|
3QF6
 
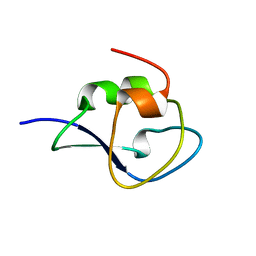 | | Neutron structure of type-III Antifreeze Protein allows the reconstruction of AFP-ice interface | | Descriptor: | Type-3 ice-structuring protein HPLC 12 | | Authors: | Howard, E.I, Blakeley, M.P, Haertlein, M, Petit-Haertlein, I, Mitschler, A, Fisher, S.J, Cousido-Siah, A, Salvay, A.G, Popov, A, Muller-Dieckmann, C, Petrova, T, Podjarny, A. | | Deposit date: | 2011-01-21 | | Release date: | 2011-06-22 | | Last modified: | 2024-03-20 | | Method: | NEUTRON DIFFRACTION (1.85 Å) | | Cite: | Neutron structure of type-III antifreeze protein allows the reconstruction of AFP-ice interface.
J.Mol.Recognit., 24, 2011
|
|
3QPL
 
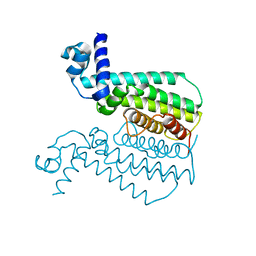 | | G106W mutant of EthR from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Willery, E, Hoos, S, Lecat-Guillet, N, Frenois, F, Dirie, B, Villeret, V, England, P, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-02-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
4KJK
 
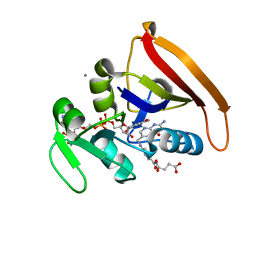 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KQ2
 
 | | Glucose1,2cyclic phosphate bound activated state of Yeast Glycogen Synthase | | Descriptor: | (2R,3aR,5R,6S,7S,7aR)-5-(hydroxymethyl)tetrahydro-3aH-[1,3,2]dioxaphospholo[4,5-b]pyran-2,6,7-triol 2-oxide, 6-O-phosphono-alpha-D-glucopyranose, BARIUM ION, ... | | Authors: | Chikwana, V.M, Hurley, T.D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for 2'-phosphate incorporation into glycogen by glycogen synthase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3OY6
 
 | |
3PDO
 
 | | Crystal Structure of HLA-DR1 with CLIP102-120 | | Descriptor: | FORMIC ACID, GLYCEROL, HLA class II histocompatibility antigen gamma chain, ... | | Authors: | Gunther, S, Schlundt, A, Sticht, J, Roske, Y, Heinemann, U, Wiesmuller, K.-H, Jung, G, Falk, K, Rotzschke, O, Freund, C. | | Deposit date: | 2010-10-23 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Bidirectional binding of invariant chain peptides to an MHC class II molecule.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3PLC
 
 | | Crystal structure of Beta-Cardiotoxin, a novel three-finger cardiotoxin from the venom of Ophiophagus hannah | | Descriptor: | Beta-cardiotoxin OH-27 | | Authors: | Roy, A, Qingxiang, S, Kini, R.M, Sivaraman, J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-11-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Identification of a alpha-helical molten globule intermediate and structural characterization of beta-cardiotoxin, an all beta-sheet protein isolated from the venom of Ophiophagus hannah (king cobra).
Protein Sci., 28, 2019
|
|
3PF4
 
 | | Crystal structure of Bs-CspB in complex with r(GUCUUUA) | | Descriptor: | Cold shock protein cspB, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Sachs, R, Max, K.E.A, Heinemann, U. | | Deposit date: | 2010-10-27 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | RNA single strands bind to a conserved surface of the major cold shock protein in crystals and solution.
Rna, 18, 2012
|
|
3PF5
 
 | |
3PFU
 
 | | N-terminal domain of Thiol:disulfide interchange protein DsbD in its reduced form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Thiol:disulfide interchange protein dsbD | | Authors: | Mavridou, D.A.I, Saridakis, E, Ferguson, S.J, Redfield, C. | | Deposit date: | 2010-10-29 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxidation state-dependent protein-protein interactions in disulfide cascades
J.Biol.Chem., 286, 2011
|
|
4I9H
 
 | | Crystal structure of rabbit LDHA in complex with AP28669 | | Descriptor: | 1-O-[3-(5-carboxypyridin-2-yl)-5-fluorophenyl]-6-O-[4-({[(5-carboxypyridin-2-yl)sulfanyl]acetyl}amino)-2-chloro-5-methoxyphenyl]-D-mannitol, L-lactate dehydrogenase A chain | | Authors: | Zhou, T, Stephan, Z.G, Kohlmann, A, Li, F, Commodore, L, Greenfield, M.T, Shakespeare, W.C, Zhu, X, Dalgarno, D.C. | | Deposit date: | 2012-12-05 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Fragment growing and linking lead to novel nanomolar lactate dehydrogenase inhibitors.
J.Med.Chem., 56, 2013
|
|
3QLP
 
 | | X-ray structure of the complex between human alpha thrombin and a modified thrombin binding aptamer (mTBA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, POTASSIUM ION, ... | | Authors: | Russo Krauss, I, Merlino, A, Mazzarella, L, Sica, F. | | Deposit date: | 2011-02-03 | | Release date: | 2011-10-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thrombin-aptamer recognition: a revealed ambiguity.
Nucleic Acids Res., 39, 2011
|
|
3QX9
 
 | | Crystal structure of MID domain from hAGO2 in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein argonaute-2 | | Authors: | Frank, F, Fabian, M.R, Stepinski, J, Jemielity, J, Darzynkiewicz, E, Sonenberg, N, Nagar, B. | | Deposit date: | 2011-03-01 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of 5'-mRNA-cap interactions with the human AGO2 MID domain.
Embo Rep., 12, 2011
|
|
4KUP
 
 | | Endothiapepsin in complex with 20mM acylhydrazone inhibitor | | Descriptor: | (2S)-2-azanyl-3-(3H-indol-3-yl)-N-[(E)-(2,4,6-trimethylphenyl)methylideneamino]propanamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2013-05-22 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.311 Å) | | Cite: | Structure-based design of inhibitors of the aspartic protease endothiapepsin by exploiting dynamic combinatorial chemistry.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
3QXR
 
 | |
4L6C
 
 | | Crystal structure of human mitochondrial deoxyribonucleotidase in complex with the inhibitor pib-t | | Descriptor: | 1,2-ETHANEDIOL, 1-{2-deoxy-3,5-O-[(4-iodophenyl)(phosphono)methylidene]-beta-D-threo-pentofuranosyl}-5-methylpyrimidine-2,4(1H,3H)-dione, 5'(3')-deoxyribonucleotidase, ... | | Authors: | Pachl, P, Brynda, J, Rezacova, P. | | Deposit date: | 2013-06-12 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conformationally constrained nucleoside phosphonic acids - potent inhibitors of human mitochondrial and cytosolic 5'(3')-nucleotidases.
Org.Biomol.Chem., 12, 2014
|
|
4L9U
 
 | | Structure of C-terminal coiled coil of RasGRP1 | | Descriptor: | GLYCEROL, RAS guanyl-releasing protein 1, SULFATE ION | | Authors: | Iwig, J.S, Vercoulen, Y, Das, R, Barros, T, Limnander, A, Che, Y, Pelton, J.G, Wemmer, D.E, Roose, J.P, Kuriyan, J. | | Deposit date: | 2013-06-18 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6014 Å) | | Cite: | Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1.
Elife, 2, 2013
|
|
5IB2
 
 | | Crystal structure of HLA-B*27:05 complexed with the self-peptide pVIPR | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
5IB5
 
 | | Crystal structure of HLA-B*27:09 complexed with the self-peptide pVIPR and Copper | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Janke, R, Ballaschk, M, Schmieder, P, Uchanska-Ziegler, B, Ziegler, A, Loll, B. | | Deposit date: | 2016-02-22 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Metal-triggered conformational reorientation of a self-peptide bound to a disease-associated HLA-B*27 subtype.
J.Biol.Chem., 2019
|
|
6M8N
 
 | | Endo-fucoidan hydrolase P5AFcnA from glycoside hydrolase family 107 | | Descriptor: | CALCIUM ION, MALONATE ION, P5AFcnA | | Authors: | Boraston, A.B, Vickers, C.J, Abe, K, Salama-Alber, O. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to alpha-l-fucosidases from GH29.
J. Biol. Chem., 293, 2018
|
|
