6BA2
 
 | | Crystal structure of MYST acetyltransferase domain in complex with inhibitor | | Descriptor: | 4-fluoro-5-methyl-N'-(phenylsulfonyl)[1,1'-biphenyl]-3-carbohydrazide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Hermans, S.J, Chung, M.C, Peat, T.S, Baell, J.B, Thomas, T, Parker, M.W. | | Deposit date: | 2017-10-11 | | Release date: | 2018-08-01 | | Last modified: | 2020-01-08 | | Method: | X-RAY DIFFRACTION (1.85003817 Å) | | Cite: | Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth.
Nature, 560, 2018
|
|
3CVW
 
 | | Drosophila melanogaster (6-4) photolyase H365N mutant bound to ds DNA with a T-T (6-4) photolesion and cofactor F0 | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a cofactor f0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
6BAA
 
 | | Cryo-EM structure of the pancreatic beta-cell KATP channel bound to ATP and glibenclamide | | Descriptor: | 5-chloro-N-(2-{4-[(cyclohexylcarbamoyl)sulfamoyl]phenyl}ethyl)-2-methoxybenzamide, ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ... | | Authors: | Martin, G.M, Yoshioka, C, Shyng, S.L. | | Deposit date: | 2017-10-12 | | Release date: | 2017-11-01 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Anti-diabetic drug binding site in a mammalian KATPchannel revealed by Cryo-EM.
Elife, 6, 2017
|
|
3D0G
 
 | |
3D2N
 
 | |
6ATK
 
 | |
6AUY
 
 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 4-Methyl-6-(3-(3-(methylamino)propyl)phenethyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-(2-{3-[3-(methylamino)propyl]phenyl}ethyl)pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
6AVE
 
 | |
6AVT
 
 | | STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) TERNARY COMPLEX WITH A DOUBLE STRANDED DNA AND AN INCOMING D4TTP AT PH 9.5 WITH CROSS-LINKING TO FIRST BASE TEMPLATE OVERHANG | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, DNA (27-MER), DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*(DDG))-3'), ... | | Authors: | Martinez, S.E, Das, K, Arnold, E. | | Deposit date: | 2017-09-04 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.603 Å) | | Cite: | Structure of HIV-1 reverse transcriptase/d4TTP complex: Novel DNA cross-linking site and pH-dependent conformational changes.
Protein Sci., 28, 2019
|
|
6AT0
 
 | |
3CHP
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid | | Descriptor: | (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHY
 
 | |
6CXJ
 
 | |
6D07
 
 | |
3CK7
 
 | | B. thetaiotaomicron SusD with alpha-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclohexakis-(1-4)-(alpha-D-glucopyranose), SusD | | Authors: | Koropatkin, N.M, Martens, E.C, Gordon, J.I, Smith, T.J. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices.
Structure, 16, 2008
|
|
6CVM
 
 | | Atomic resolution cryo-EM structure of beta-galactosidase | | Descriptor: | 2-phenylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, MAGNESIUM ION, ... | | Authors: | Subramaniam, S, Bartesaghi, A, Banerjee, S, Zhu, X, Milne, J.L.S. | | Deposit date: | 2018-03-28 | | Release date: | 2018-05-30 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Atomic Resolution Cryo-EM Structure of beta-Galactosidase.
Structure, 26, 2018
|
|
3COA
 
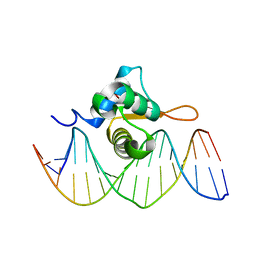 | | Crystal Structure of FoxO1 DBD Bound to IRE DNA | | Descriptor: | CALCIUM ION, DNA (5'-D(*DCP*DAP*DAP*DGP*DCP*DAP*DAP*DAP*DAP*DCP*DAP*DAP*DAP*DCP*DCP*DA)-3'), DNA (5'-D(*DTP*DGP*DGP*DTP*DTP*DTP*DGP*DTP*DTP*DTP*DTP*DGP*DCP*DTP*DTP*DG)-3'), ... | | Authors: | Brent, M.M, Anand, R, Marmorstein, R. | | Deposit date: | 2008-03-27 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for DNA Recognition by FoxO1 and Its Regulation by Posttranslational Modification.
Structure, 16, 2008
|
|
3CRU
 
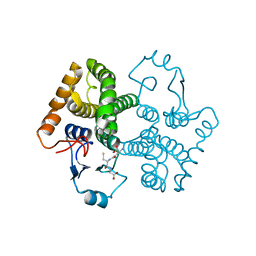 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
3CSH
 
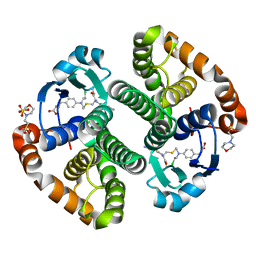 | |
3CQK
 
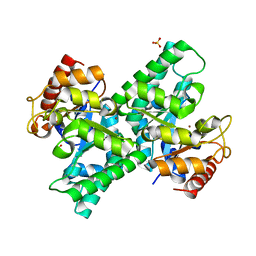 | | Crystal Structure of L-xylulose-5-phosphate 3-epimerase UlaE (form B) complex with Zn2+ and sulfate | | Descriptor: | L-ribulose-5-phosphate 3-epimerase ulaE, SULFATE ION, ZINC ION | | Authors: | Shi, R, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2008-04-03 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structure of L-xylulose-5-Phosphate 3-epimerase (UlaE) from the anaerobic L-ascorbate utilization pathway of Escherichia coli: identification of a novel phosphate binding motif within a TIM barrel fold.
J.Bacteriol., 190, 2008
|
|
6D3H
 
 | | FT_T dioxygenase with bound dichlorprop | | Descriptor: | (2R)-2-(2,4-dichlorophenoxy)propanoic acid, 2-OXOGLUTARIC ACID, CHLORIDE ION, ... | | Authors: | Rydel, T.J, Halls, C.E. | | Deposit date: | 2018-04-16 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
3CRT
 
 | | Structural characterization of an engineered allosteric protein | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme | | Authors: | Sagermann, M, Chapleau, R, DeLorimier, E, Lei, M. | | Deposit date: | 2008-04-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Using affinity chromatography to engineer and characterize pH-dependent protein switches.
Protein Sci., 18, 2009
|
|
6D0O
 
 | | rdpA dioxygenase holoenzyme | | Descriptor: | (R)-phenoxypropionate/alpha-ketoglutarate-dioxygenase, 2-OXOGLUTARIC ACID, COBALT (II) ION | | Authors: | Rydel, T.J, Sturman, E.J, Zheng, M, Evdokimov, A. | | Deposit date: | 2018-04-10 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development of enzymes for robust aryloxyphenoxypropionate and synthetic auxin herbicide tolerance traits in maize and soybean crops.
Pest Manag. Sci., 75, 2019
|
|
3CVX
 
 | | Drosophila melanogaster (6-4) photolyase H369M mutant bound to ds DNA with a T-T (6-4) photolesion | | Descriptor: | DNA (5'-D(*DAP*DCP*DAP*DGP*DCP*DGP*DGP*(64T)P*(5PY)P*DGP*DCP*DAP*DGP*DGP*DT)-3'), DNA (5'-D(*DTP*DAP*DCP*DCP*DTP*DGP*DCP*DAP*DAP*DCP*DCP*DGP*DCP*DTP*DG)-3'), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maul, M.J, Barends, T.R.M, Glas, A.F, Cryle, M.J, Schlichting, I, Carell, T. | | Deposit date: | 2008-04-20 | | Release date: | 2009-10-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and mechanism of a coenzyme F0
accelerated (6-4) photolyase from the fruit fly
To be Published
|
|
6CX0
 
 | | Structure of AtTPC1 D376A | | Descriptor: | (1S,3R)-1-(3-{[4-(2-fluorophenyl)piperazin-1-yl]methyl}-4-methoxyphenyl)-2,3,4,9-tetrahydro-1H-beta-carboline-3-carboxylic acid, CALCIUM ION, Two pore calcium channel protein 1 | | Authors: | Kintzer, A.F, Stroud, R.M. | | Deposit date: | 2018-04-02 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural basis for activation of voltage sensor domains in an ion channel TPC1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
