7U92
 
 | | SARS-CoV-2 Main Protease (Mpro) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-03-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUG
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with ML1006a | | Descriptor: | (1R,2S,5S)-N-{(2S,3R)-4-(azetidin-1-yl)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
7UUP
 
 | | SARS-CoV-2 Main Protease S144A (Mpro S144A) in Complex with Nirmatrelvir (PF-07321332) | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, CHLORIDE ION | | Authors: | Westberg, M, Fernandez, D, Lin, M.Z. | | Deposit date: | 2022-04-28 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An orally bioavailable SARS-CoV-2 main protease inhibitor exhibits improved affinity and reduced sensitivity to mutations.
Sci Transl Med, 16, 2024
|
|
6BQK
 
 | |
5HU1
 
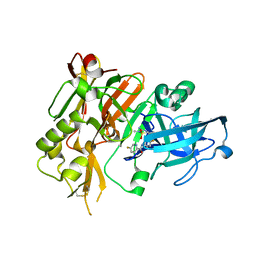 | | BACE1 in complex with (R)-N-(3-(3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl)-4-fluorophenyl)-5-fluoropicolinamide | | Descriptor: | Beta-secretase 1, N-{3-[(5R)-3-amino-2,5-dimethyl-1,1-dioxido-5,6-dihydro-2H-1,2,4-thiadiazin-5-yl]-4-fluorophenyl}-5-fluoropyridine-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
5HU0
 
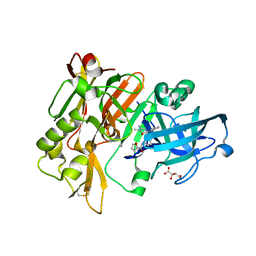 | | BACE1 in complex with 4-(3-(furan-2-carboxamido)phenyl)-1-methyl-5-oxo-4-phenylimidazolidin-2-iminium | | Descriptor: | Beta-secretase 1, L(+)-TARTARIC ACID, N-{3-[(2E,4R)-2-imino-1-methyl-5-oxo-4-phenylimidazolidin-4-yl]phenyl}furan-2-carboxamide | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
5HTZ
 
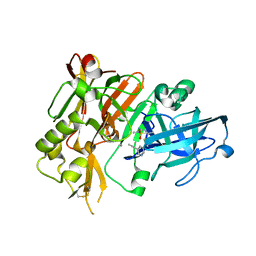 | | BACE1 in complex with (S)-5-(3-chloro-5-(5-(prop-1-yn-1-yl)pyridin-3-yl)thiophen-2-yl)-2,5-dimethyl-1,2,4-thiadiazinan-3-iminium 1,1-dioxide | | Descriptor: | (3E,5S)-5-{3-chloro-5-[5-(prop-1-yn-1-yl)pyridin-3-yl]thiophen-2-yl}-2,5-dimethyl-1,2,4-thiadiazinan-3-imine 1,1-dioxide, Beta-secretase 1 | | Authors: | Orth, P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-11-09 | | Last modified: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of the 3-Imino-1,2,4-thiadiazinane 1,1-Dioxide Derivative Verubecestat (MK-8931)-A beta-Site Amyloid Precursor Protein Cleaving Enzyme 1 Inhibitor for the Treatment of Alzheimer's Disease.
J. Med. Chem., 59, 2016
|
|
7E2E
 
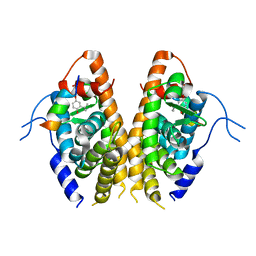 | | Crystal structure of the Estrogen-Related Receptor alpha (ERRalpha) ligand-binding domain (LBD) in complex with an agonist DS45500853 and a PGC-1alpha peptide | | Descriptor: | 1-[4-(3-tert-butyl-4-oxidanyl-phenoxy)phenyl]ethanone, IODIDE ION, Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, ... | | Authors: | Ito, S, Shinozuka, T, Kimura, T, Izumi, M, Wakabayashi, K. | | Deposit date: | 2021-02-05 | | Release date: | 2021-06-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a Novel Class of ERR alpha Agonists.
Acs Med.Chem.Lett., 12, 2021
|
|
6QD8
 
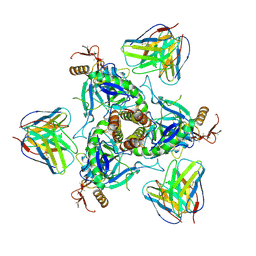 | | EM structure of a EBOV-GP bound to 4M0368 neutralizing antibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, Envelope glycoprotein,Virion spike glycoprotein,EBOV-GP1, ... | | Authors: | Diskin, R, Cohen-Dvashi, H. | | Deposit date: | 2019-01-01 | | Release date: | 2019-10-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | rVSV-ZEBOV induces a polyclonal and convergent B cell response with potent Ebola virus-neutralizing antibodies
Nat.Med. (N.Y.), 2019
|
|
8SG2
 
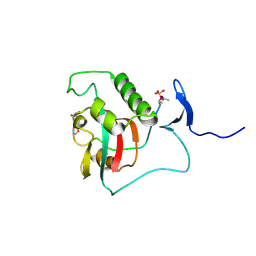 | | BIVALENT INTERACTIONS OF PIN1 WITH THE C-TERMINAL TAIL OF PKC | | Descriptor: | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, Protein kinase C beta type | | Authors: | Dixit, K, Yang, Y, Chen, X.R, Igumenova, T.I. | | Deposit date: | 2023-04-11 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel bivalent interaction mode underlies a non-catalytic mechanism for Pin1-mediated protein kinase C regulation.
Elife, 13, 2024
|
|
6W2T
 
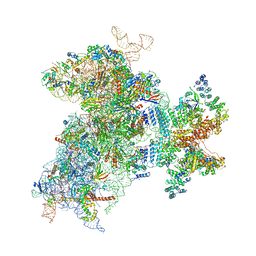 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the closed state (Class 2) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
6W2S
 
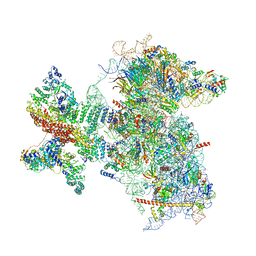 | | Structure of the Cricket Paralysis Virus 5-UTR IRES (CrPV 5-UTR-IRES) bound to the small ribosomal subunit in the open state (Class 1) | | Descriptor: | 18S rRNA, CrPV 5'-UTR IRES, Eukaryotic translation initiation factor 3 subunit A, ... | | Authors: | Neupane, R, Pisareva, V, Rodriguez, C.F, Pisarev, A, Fernandez, I.S. | | Deposit date: | 2020-03-08 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | A complex IRES at the 5'-UTR of a viral mRNA assembles a functional 48S complex via an uAUG intermediate.
Elife, 9, 2020
|
|
7QVP
 
 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
5G4S
 
 | | BROMODOMAIN OF HUMAN BRPF1 WITH N-1,3-dimethyl-6-2R-2- methylpiperazin-1-yl-2-oxo-2,3-dihydro-1H-1,3-benzodiazol-5-yl-N- ethyl-2-methoxybenzamide | | Descriptor: | 1,2-ETHANEDIOL, N-[1,3-dimethyl-6-[(2R)-2-methylpiperazin-1-yl]-2-oxidanylidene-benzimidazol-5-yl]-N-ethyl-2-methoxy-benzamide, PEREGRIN | | Authors: | Chung, C. | | Deposit date: | 2016-05-16 | | Release date: | 2016-07-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Gsk6853, a Chemical Probe for Inhibition of the Brpf1 Bromodomain.
Acs Med.Chem.Lett., 7, 2016
|
|
7SLS
 
 | | HIV Reverse Transcriptase with compound Pyr02 | | Descriptor: | 5-(difluoromethyl)-3-{[1-{[(3S)-5-fluoro-2-methyl-6-oxo-3,6-dihydropyridin-3-yl]methyl}-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl]oxy}-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.078 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
7SLR
 
 | | HIV Reverse Transcriptase with compound Pyr01 | | Descriptor: | 5-(difluoromethyl)-3-({1-[(5-fluoro-2-oxo-1,2-dihydropyridin-3-yl)methyl]-6-oxo-4-(1,1,2,2-tetrafluoroethyl)-1,6-dihydropyrimidin-5-yl}oxy)-2-methylbenzonitrile, Reverse transcriptase/ribonuclease H | | Authors: | Klein, D.J, Zebisch, M, Gu, M. | | Deposit date: | 2021-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.179 Å) | | Cite: | Potent targeted activator of cell kill molecules eliminate cells expressing HIV-1.
Sci Transl Med, 15, 2023
|
|
8G2U
 
 | | Time-resolved cryo-EM study of the 70S recycling by the HflX:control-apo-70S at 900ms | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Bhattacharjee, S, Brown, P.Z, Frank, J. | | Deposit date: | 2023-02-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Time resolution in cryo-EM using a PDMS-based microfluidic chip assembly and its application to the study of HflX-mediated ribosome recycling.
Cell, 187, 2024
|
|
8TDQ
 
 | | SFX-XFEL structure of CYP121 cocrystallized with substrate cYY | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
8TDP
 
 | | Time-resolved SFX-XFEL crystal structure of CYP121 bound with cYY reacted with peracetic acid for 200 milliseconds | | Descriptor: | (3S,6S)-3,6-bis(4-hydroxybenzyl)piperazine-2,5-dione, HYDROGEN PEROXIDE, Mycocyclosin synthase, ... | | Authors: | Nguyen, R.C, Dasgupta, M, Bhowmick, A, Kern, J.F, Liu, A. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | In Situ Structural Observation of a Substrate- and Peroxide-Bound High-Spin Ferric-Hydroperoxo Intermediate in the P450 Enzyme CYP121.
J.Am.Chem.Soc., 145, 2023
|
|
6BQJ
 
 | |
5FP7
 
 | |
7UOV
 
 | |
7UOT
 
 | | Native Lassa glycoprotein in complex with neutralizing antibodies 8.9F and 37.2D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 37.2D heavy chain (variable domain), ... | | Authors: | Li, H, Saphire, E.O. | | Deposit date: | 2022-04-13 | | Release date: | 2022-11-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | A cocktail of protective antibodies subverts the dense glycan shield of Lassa virus.
Sci Transl Med, 14, 2022
|
|
3VEK
 
 | | Both Zn Fingers of GATA1 Bound to Palindromic DNA Recognition Site, P1 Crystal Form | | Descriptor: | DNA (5'-D(*AP*AP*GP*AP*GP*TP*CP*CP*AP*TP*CP*TP*GP*AP*TP*AP*AP*GP*AP*C)-3'), DNA (5'-D(*TP*TP*GP*TP*CP*TP*TP*AP*TP*CP*AP*GP*AP*TP*GP*GP*AP*CP*TP*C)-3'), Erythroid transcription factor, ... | | Authors: | Jacques, D.A, Ripin, N, Wilkinson-White, L.E, Guss, J.M, Matthews, J.M. | | Deposit date: | 2012-01-09 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | GATA1 directly mediates interactions with closely spaced pseudopalindromic but not distantly spaced double GATA sites on DNA.
Protein Sci., 24, 2015
|
|
7C2W
 
 | |
