6C0N
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with non-nucleoside inhibitor 25a | | Descriptor: | 1,2-ETHANEDIOL, 4-({4-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}thieno[3,2-d]pyrimidin-2-yl)amino]piperidin-1-yl}methyl)benzene-1-sulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Yang, Y, Nguyen, L.A, Smithline, Z.B, Steitz, T.A. | | Deposit date: | 2018-01-01 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Structural basis for potent and broad inhibition of HIV-1 RT by thiophene[3,2-d]pyrimidine non-nucleoside inhibitors.
Elife, 7, 2018
|
|
6C1G
 
 | | High-Resolution Cryo-EM Structures of Actin-bound Myosin States Reveal the Mechanism of Myosin Force Sensing | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Mentes, A, Huehn, A, Liu, X, Zwolak, A, Dominguez, R, Shuman, H, Ostap, E.M, Sindelar, C.V. | | Deposit date: | 2018-01-04 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | High-resolution cryo-EM structures of actin-bound myosin states reveal the mechanism of myosin force sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3DI6
 
 | | HIV-1 RT with pyridazinone non-nucleoside inhibitor | | Descriptor: | 6-(4-chloro-2-fluoro-3-phenoxybenzyl)pyridazin-3(2H)-one, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Harris, S.F, Villasenor, A, Dunten, P. | | Deposit date: | 2008-06-19 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery and optimization of pyridazinone non-nucleoside inhibitors of HIV-1 reverse transcriptase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3DQS
 
 | | Structure of endothelial NOS heme domain in complex with a inhibitor (+-)-N1-{cis-4'-[(6"-amino-4"-methylpyridin-2"-yl)methyl]pyrrolidin-3'-yl}-N2-(4'-chlorobenzyl)ethane-1,2-diamine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Igarashi, J, Li, H, Poulos, T.L. | | Deposit date: | 2008-07-09 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal structures of constitutive nitric oxide synthases in complex with de novo designed inhibitors.
J.Med.Chem., 52, 2009
|
|
6BNO
 
 | | Structure of bare actin filament | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Gurel, P.S, Alushin, G.A. | | Deposit date: | 2017-11-17 | | Release date: | 2018-01-10 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structures reveal specialization at the myosin VI-actin interface and a mechanism of force sensitivity.
Elife, 6, 2017
|
|
3DRR
 
 | | HIV reverse transcriptase Y181C mutant in complex with inhibitor R8e | | Descriptor: | 3-{5-[(6-amino-1H-pyrazolo[3,4-b]pyridin-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Yan, Y. | | Deposit date: | 2008-07-11 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of 3-{5-[(6-Amino-1H-pyrazolo[3,4-b]pyridine-3-yl)methoxy]-2-chlorophenoxy}-5-chlorobenzonitrile (MK-4965): A Potent, Orally Bioavailable HIV-1 Non-Nucleoside Reverse Transcriptase Inhibitor with Improved Potency against Key Mutant Viruses.
J.Med.Chem., 51, 2008
|
|
3DTU
 
 | | Catalytic core subunits (I and II) of cytochrome c oxidase from Rhodobacter sphaeroides complexed with deoxycholic acid | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, CADMIUM ION, CALCIUM ION, ... | | Authors: | Qin, L, Mills, D.A, Buhrow, L, Hiser, C, Ferguson-Miller, S. | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A conserved steroid binding site in cytochrome C oxidase.
Biochemistry, 47, 2008
|
|
6CA4
 
 | | Crystal structure of humanized D. rerio TDP2 by 14 mutations | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Shi, K, Aihrara, H. | | Deposit date: | 2018-01-29 | | Release date: | 2018-05-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.623 Å) | | Cite: | New fluorescence-based high-throughput screening assay for small molecule inhibitors of tyrosyl-DNA phosphodiesterase 2 (TDP2).
Eur J Pharm Sci, 118, 2018
|
|
3DUT
 
 | | The high salt (phosphate) crystal structure of deoxy hemoglobin E (GLU26LYS) at physiological pH (pH 7.35) | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PHOSPHATE ION, ... | | Authors: | Malashkevich, V.N, Balazs, T.C, Almo, S.C, Hirsch, R.E. | | Deposit date: | 2008-07-17 | | Release date: | 2009-08-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The high salt (phosphate) crystal structure of deoxy
hemoglobin E (GLU26LYS) at physiological pH (pH 7.35)
To be Published
|
|
6BK0
 
 | |
6BSG
 
 | | Structure of HIV-1 RT complexed with RNA/DNA hybrid in an RNA hydrolysis-off mode | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, CALCIUM ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tian, L, Kim, M, Yang, W. | | Deposit date: | 2017-12-03 | | Release date: | 2018-01-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3DYE
 
 | | Crystal structure of AED7-norepineprhine complex | | Descriptor: | BROMIDE ION, D7 protein, GLYCEROL, ... | | Authors: | Andersen, J.F, Calvo, E, Mans, B.J, Ribeiro, J.M. | | Deposit date: | 2008-07-27 | | Release date: | 2009-02-03 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DYM
 
 | | E. coli (lacZ) beta-galactosidase (H418E) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Juers, D.H, Huber, R.E, Matthews, B.W. | | Deposit date: | 2008-07-28 | | Release date: | 2008-10-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Direct and indirect roles of His-418 in metal binding and in the activity of beta-galactosidase (E. coli).
Protein Sci., 18, 2009
|
|
6CIK
 
 | | Pre-Reaction Complex, RAG1(E962Q)/2-intact/nicked 12/23RSS complex in Mn2+ | | Descriptor: | DNA (5'-D(*AP*TP*CP*TP*GP*GP*CP*CP*TP*GP*TP*CP*TP*TP*A)-3'), High mobility group protein B1, Intact 12RSS substrate forward strand, ... | | Authors: | Chuenchor, W, Chen, X, Kim, M.S, Gellert, M, Yang, W. | | Deposit date: | 2018-02-24 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Cracking the DNA Code for V(D)J Recombination.
Mol. Cell, 70, 2018
|
|
3EE2
 
 | |
6CO8
 
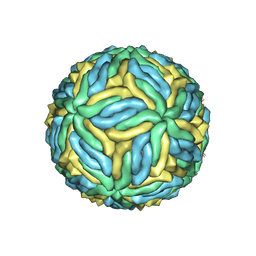 | | Structure of Zika virus at a resolution of 3.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, E protein, M protein | | Authors: | Sevvana, M, Long, F, Miller, A.J, Klose, T, Buda, G, Sun, L, Kuhn, R.J, Rossmann, M.R. | | Deposit date: | 2018-03-12 | | Release date: | 2018-07-04 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Refinement and Analysis of the Mature Zika Virus Cryo-EM Structure at 3.1 angstrom Resolution.
Structure, 26, 2018
|
|
3EH5
 
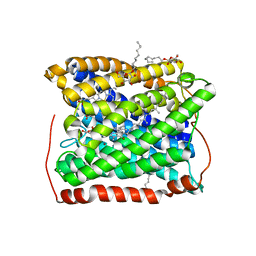 | | Structure of the reduced form of cytochrome ba3 oxidase from Thermus thermophilus | | Descriptor: | COPPER (I) ION, Cytochrome c oxidase polypeptide 2A, Cytochrome c oxidase subunit 1, ... | | Authors: | Liu, B, Chen, Y, Doukov, T, Soltis, S.M, Stout, D, Fee, J.A. | | Deposit date: | 2008-09-11 | | Release date: | 2009-04-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme.
Biochemistry, 48, 2009
|
|
3EPX
 
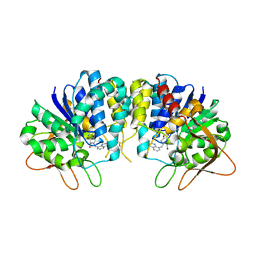 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase in complex with the inhibitor (2R,3R,4S)-2-(hydroxymethyl)-1-(quinolin-8-ylmethyl)pyrrolidin-3,4-diol | | Descriptor: | (2R,3R,4S)-2-(hydroxymethyl)-1-(quinolin-8-ylmethyl)pyrrolidine-3,4-diol, CALCIUM ION, GLYCEROL, ... | | Authors: | Versees, W, Goeminne, A, Berg, M, Vandemeulebroucke, A, Haemers, A, Augustyns, K, Steyaert, J. | | Deposit date: | 2008-09-30 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of T. vivax nucleoside hydrolase in complex with new potent and specific inhibitors.
Biochim.Biophys.Acta, 1794, 2009
|
|
6CQI
 
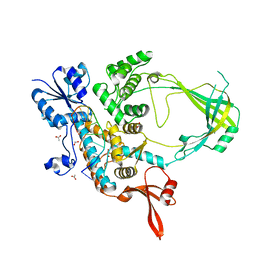 | | 2.42A Crystal structure of Mycobacterium tuberculosis Topoisomerase I in complex with an oligonucleotide MTS2-11 | | Descriptor: | ACETATE ION, DNA (5'-D(P*TP*TP*CP*CP*GP*CP*TP*TP*GP*A)-3'), DNA topoisomerase 1, ... | | Authors: | Cao, N, Thirunavukkarasu, A, Tan, K, Tse-Dinh, Y.-C. | | Deposit date: | 2018-03-15 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Investigating mycobacterial topoisomerase I mechanism from the analysis of metal and DNA substrate interactions at the active site.
Nucleic Acids Res., 46, 2018
|
|
6CB2
 
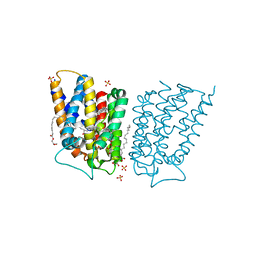 | | Crystal structure of Escherichia coli UppP | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, SULFATE ION, Undecaprenyl-diphosphatase | | Authors: | Workman, S.D, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2018-02-01 | | Release date: | 2018-03-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of an intramembranal phosphatase central to bacterial cell-wall peptidoglycan biosynthesis and lipid recycling.
Nat Commun, 9, 2018
|
|
6CGL
 
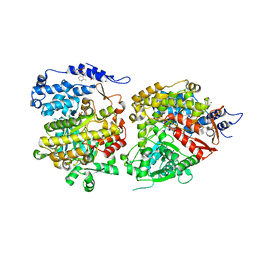 | |
6CHM
 
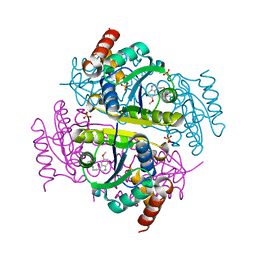 | |
6CBV
 
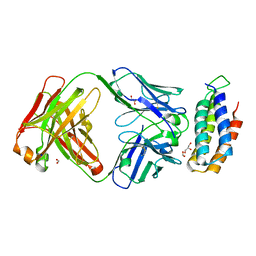 | | Crystal structure of BRIL bound to an affinity matured synthetic antibody. | | Descriptor: | BRIL, FORMIC ACID, GLYCEROL, ... | | Authors: | Mukherjee, S, Skrobek, B, Kossiakoff, A.A. | | Deposit date: | 2018-02-05 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.872 Å) | | Cite: | Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Nat Commun, 11, 2020
|
|
6CCK
 
 | | Crystal structure of E.coli Phosphopantetheine Adenylyltransferase (PPAT/CoaD) in complex with (R)-3-(3-chlorophenyl)-3-((5-methyl-7-oxo-4,7-dihydro-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino)propanenitrile | | Descriptor: | (3R)-3-(3-chlorophenyl)-3-[(5-methyl-7-oxo-6,7-dihydro[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)amino]propanenitrile, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-02-07 | | Release date: | 2018-03-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Fragment-Based Drug Discovery of Inhibitors of Phosphopantetheine Adenylyltransferase from Gram-Negative Bacteria.
J. Med. Chem., 61, 2018
|
|
3E32
 
 | | Protein farnesyltransferase complexed with FPP and ethylenediamine scaffold inhibitor 2 | | Descriptor: | FARNESYL DIPHOSPHATE, N-benzyl-N-(2-{(4-cyanophenyl)[(1-methyl-1H-imidazol-5-yl)methyl]amino}ethyl)-1-methyl-1H-imidazole-4-sulfonamide, Protein farnesyltransferase subunit beta, ... | | Authors: | Hast, M.A, Beese, L.S. | | Deposit date: | 2008-08-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for binding and selectivity of antimalarial and anticancer ethylenediamine inhibitors to protein farnesyltransferase.
Chem.Biol., 16, 2009
|
|
