3BLT
 
 | |
6A95
 
 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with tetrodotoxin and Dc1a | | Descriptor: | (1R,5R,6R,7R,9S,11S,12S,13S,14S)-3-amino-14-(hydroxymethyl)-8,10-dioxa-2,4-diazatetracyclo[7.3.1.1~7,11~.0~1,6~]tetradec-3-ene-5,9,12,13,14-pentol (non-preferred name), 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
3BNY
 
 | | Crystal structure of aristolochene synthase complexed with 2-fluorofarnesyl diphosphate (2F-FPP) | | Descriptor: | (2Z,6E)-2-fluoro-3,7,11-trimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Christianson, D.W. | | Deposit date: | 2007-12-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis
J.Biol.Chem., 283, 2008
|
|
6A3F
 
 | | Levoglucosan dehydrogenase, apo form | | Descriptor: | Putative dehydrogenase, SULFATE ION | | Authors: | Sugiura, M, Yamada, C, Arakawa, T, Fushinobu, S. | | Deposit date: | 2018-06-15 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification, functional characterization, and crystal structure determination of bacterial levoglucosan dehydrogenase.
J. Biol. Chem., 293, 2018
|
|
6A91
 
 | | Complex of voltage-gated sodium channel NavPaS from American cockroach Periplaneta americana bound with saxitoxin and Dc1a | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shen, H.Z, li, Z.Q, Jiang, Y, Pan, X.J, Wu, J.P, Cristofori-Armstrong, B, Smith, J.J, Chin, Y.K.Y, Lei, J.L, Zhou, Q, King, G.F, Yan, N. | | Deposit date: | 2018-07-11 | | Release date: | 2018-08-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for the modulation of voltage-gated sodium channels by animal toxins.
Science, 362, 2018
|
|
6AFW
 
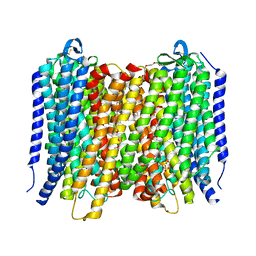 | | Proton pyrophosphatase-T228D mutant | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Tsai, J.-Y, Li, K.-M, Sun, Y.-J. | | Deposit date: | 2018-08-08 | | Release date: | 2019-04-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Roles of the Hydrophobic Gate and Exit Channel in Vigna radiata Pyrophosphatase Ion Translocation.
J. Mol. Biol., 431, 2019
|
|
6AH5
 
 | | Structure of human P2X3 receptor in complex with ATP and Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.819 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
3BYN
 
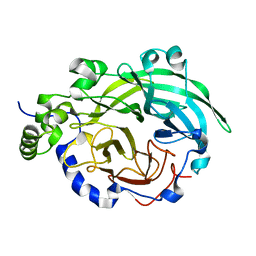 | |
3BYK
 
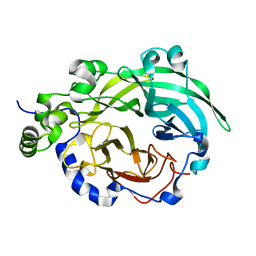 | |
3C28
 
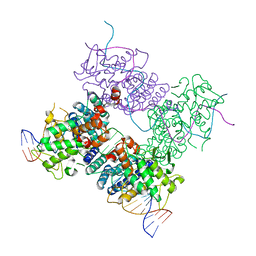 | |
6AKO
 
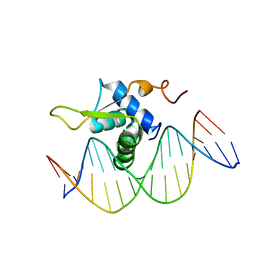 | | Crystal Structure of FOXC2 DBD Bound to DBE2 DNA | | Descriptor: | DNA (5'-D(CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Forkhead box protein C2, ... | | Authors: | Chen, X, Wei, H, Li, J, Liang, X, Dai, S, Jiang, L, Guo, M, Chen, Y. | | Deposit date: | 2018-09-03 | | Release date: | 2019-02-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Structural basis for DNA recognition by FOXC2.
Nucleic Acids Res., 47, 2019
|
|
6AK3
 
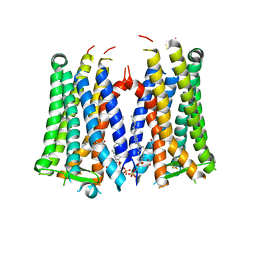 | | Crystal structure of the human prostaglandin E receptor EP3 bound to prostaglandin E2 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Prostaglandin E2 receptor EP3 subtype,Soluble cytochrome b562 | | Authors: | Morimoto, K, Suno, R, Iwata, S, Kobayashi, T. | | Deposit date: | 2018-08-29 | | Release date: | 2018-12-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the endogenous agonist-bound prostanoid receptor EP3.
Nat. Chem. Biol., 15, 2019
|
|
3BKZ
 
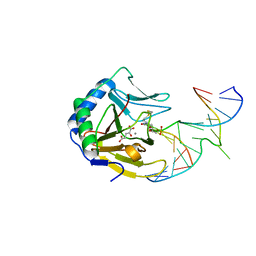 | | X-ray structure of E coli AlkB crosslinked to dsDNA in the active site | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase alkB, DNA (5'-D(*DAP*DAP*DCP*DGP*DAP*DTP*DAP*DTP*DTP*DAP*DCP*DCP*DT)-3'), ... | | Authors: | Yi, C, Yang, C.-G, He, C. | | Deposit date: | 2007-12-09 | | Release date: | 2008-04-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA
Nature, 452, 2008
|
|
3BL9
 
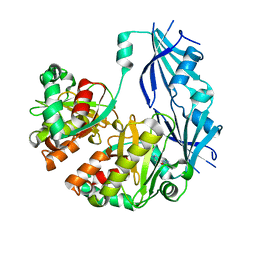 | | Synthetic Gene Encoded DcpS bound to inhibitor DG157493 | | Descriptor: | 5-{[1-(2,3-dichlorobenzyl)piperidin-4-yl]methoxy}quinazoline-2,4-diamine, Scavenger mRNA-decapping enzyme DcpS | | Authors: | Staker, B.L, Christensen, J, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-12-10 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | DcpS as a therapeutic target for spinal muscular atrophy.
Acs Chem.Biol., 3, 2008
|
|
3BSG
 
 | |
3BTF
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI. | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3DEN
 
 | | Structure of E. coli DHDPS mutant Y107W | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Pearce, F.G, Gerrard, J.A, Perugini, M.A, Jameson, G.B. | | Deposit date: | 2008-06-10 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutating the tight-dimer interface of dihydrodipicolinate synthase disrupts the enzyme quaternary structure: toward a monomeric enzyme
Biochemistry, 47, 2008
|
|
3D9G
 
 | | Nitroalkane oxidase: wild type crystallized in a trapped state forming a cyanoadduct with FAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Nitroalkane oxidase, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3DF0
 
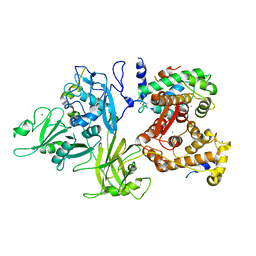 | | Calcium-dependent complex between m-calpain and calpastatin | | Descriptor: | CALCIUM ION, Calpain small subunit 1, Calpain-2 catalytic subunit, ... | | Authors: | Moldoveanu, T, Gehring, K, Green, D.R. | | Deposit date: | 2008-06-11 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Concerted multi-pronged attack by calpastatin to occlude the catalytic cleft of heterodimeric calpains.
Nature, 456, 2008
|
|
6ADQ
 
 | | Respiratory Complex CIII2CIV2SOD2 from Mycobacterium smegmatis | | Descriptor: | (2R)-2-(hexadecanoyloxy)-3-{[(S)-hydroxy{[(1R,2R,3R,4R,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propyl (9S)-9-methyloctadecanoate, (2R)-3-(((2-aminoethoxy)(hydroxy)phosphoryl)oxy)-2-(palmitoyloxy)propyl (E)-octadec-9-enoate, (2S)-1-(hexadecanoyloxy)propan-2-yl (10S)-10-methyloctadecanoate, ... | | Authors: | Gong, H.R, Xu, A, Gao, R.G, Ji, W.X, Wang, S.H, Wang, Q, Li, J, Rao, Z.H. | | Deposit date: | 2018-08-01 | | Release date: | 2018-11-14 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | An electron transfer path connects subunits of a mycobacterial respiratory supercomplex.
Science, 362, 2018
|
|
3D9F
 
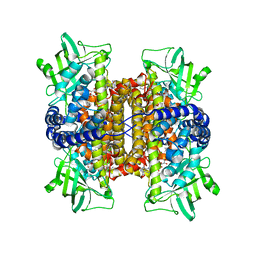 | | Nitroalkane oxidase: active site mutant S276A crystallized with 1-nitrohexane | | Descriptor: | 1-nitrohexane, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Heroux, A, Bozinovski, D.M, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2008-05-27 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of intermediates in the nitroalkane oxidase reaction.
Biochemistry, 48, 2009
|
|
3DFR
 
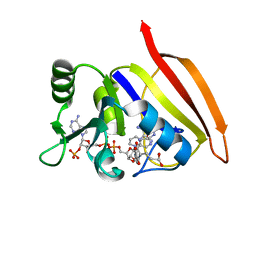 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI AND LACTOBACILLUS CASEI DIHYDROFOLATE REDUCTASE REFINED AT 1.7 ANGSTROMS RESOLUTION. I. GENERAL FEATURES AND BINDING OF METHOTREXATE | | Descriptor: | DIHYDROFOLATE REDUCTASE, METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Filman, D.J, Matthews, D.A, Bolin, J.T, Kraut, J. | | Deposit date: | 1982-06-25 | | Release date: | 1982-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of Escherichia coli and Lactobacillus casei dihydrofolate reductase refined at 1.7 A resolution. I. General features and binding of methotrexate.
J.Biol.Chem., 257, 1982
|
|
3DK8
 
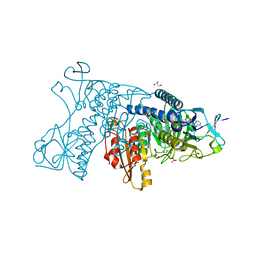 | | Catalytic cycle of human glutathione reductase near 1 A resolution | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE, GLYCEROL, ... | | Authors: | Berkholz, D.S, Faber, H.R, Savvides, S.N, Karplus, P.A. | | Deposit date: | 2008-06-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Catalytic cycle of human glutathione reductase near 1 A resolution.
J.Mol.Biol., 382, 2008
|
|
6A69
 
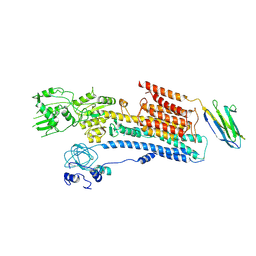 | | Cryo-EM structure of a P-type ATPase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroplastin, Plasma membrane calcium-transporting ATPase 1 | | Authors: | Gong, D.S, Chi, X.M, Ren, K, Huang, G.X.Y, Zhou, G.W, Yan, N, Lei, J.L, Zhou, Q. | | Deposit date: | 2018-06-27 | | Release date: | 2018-09-19 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin.
Nat Commun, 9, 2018
|
|
3DAW
 
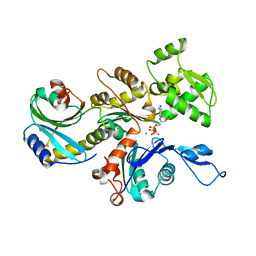 | | Structure of the actin-depolymerizing factor homology domain in complex with actin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Paavilainen, V.O, Oksanen, E, Goldman, A, Lappalainen, P. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of the actin-depolymerizing factor homology domain in complex with actin
J.Cell Biol., 182, 2008
|
|
