6Z49
 
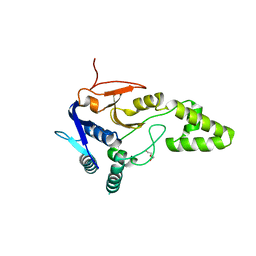 | | Crystal structure of deubiquitinase Mindy2 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-05-23 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6Z6E
 
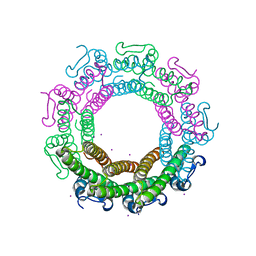 | | Crystal structure of the HK97 bacteriophage small terminase | | Descriptor: | IODIDE ION, Terminase small subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
6Z6Z
 
 | |
6Z6D
 
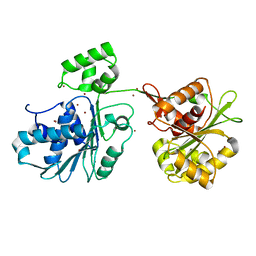 | | Crystal structure of the HK97 bacteriophage large terminase | | Descriptor: | BROMIDE ION, Terminase large subunit | | Authors: | Fung, H.K.H, Chechik, M, Baumann, C.G, Antson, A.A. | | Deposit date: | 2020-05-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of DNA packaging by a ring-type ATPase from an archetypal viral system.
Nucleic Acids Res., 50, 2022
|
|
6ZBQ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZBZ
 
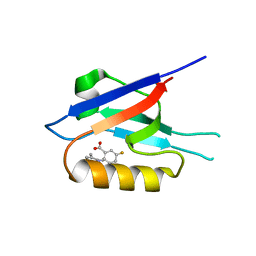 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dimethylphenyl)sulfonylamino]-5-fluoranyl-benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC7
 
 | |
1KGS
 
 | |
6ZC6
 
 | |
6ZC3
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-2-(methylsulfonylamino)benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
1KH0
 
 | | Accurate Computer Base Design of a New Backbone Conformation in the Second Turn of Protein L | | Descriptor: | protein L | | Authors: | O'Neill, J.W, Kuhlman, B, Kim, D.E, Zhang, K.Y, Baker, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-01-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Accurate computer-based design of a new backbone conformation in the second turn of protein L.
J.Mol.Biol., 315, 2002
|
|
6ZC8
 
 | |
6ZC4
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-bromanyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZFU
 
 | | Crystal structure of bovine cytochrome bc1 in complex with quinolone inhibitor RKA066 | | Descriptor: | 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ... | | Authors: | Amporndanai, K, O'Neill, P.M, Hong, W.D, Amewu, R.K, Pidathala, C, Berry, N.G, Biagini, G.A, Leung, S.C, Hasnain, S.S, Antonyuk, S.V. | | Deposit date: | 2020-06-17 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Targeting the ubiquinol-reduction (Qi) site of the mitochondrial cytochrome bc1 complex for the development of next generation quinolone antimalarials
To Be Published
|
|
1KJM
 
 | | TAP-A-associated rat MHC class I molecule | | Descriptor: | B6 Peptide, RT1 class I histocompatibility antigen, AA alpha chain, ... | | Authors: | Rudolph, M.G, Stevens, J, Speir, J.A, Trowsdale, J, Butcher, G.W, Joly, E, Wilson, I.A. | | Deposit date: | 2001-12-04 | | Release date: | 2002-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of two rat MHC class Ia (RT1-A) molecules that are associated differentially
with peptide transporter alleles TAP-A and TAP-B.
J.Mol.Biol., 324, 2002
|
|
6Z90
 
 | | Crystal structure of MINDY1 mutant-P138A | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase MINDY-1 | | Authors: | Abdul Rehman, S.A, Kulathu, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2.
Mol.Cell, 81, 2021
|
|
6ZC5
 
 | | Human Adenovirus serotype D10 FiberKnob protein | | Descriptor: | Fiber | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
6ZBK
 
 | | Crystal structure of the human complex between RPAP3 and TRBP | | Descriptor: | RISC-loading complex subunit TARBP2, RNA polymerase II-associated protein 3 | | Authors: | Charron, C, Abel, Y, Charpentier, B, Rederstorff, M. | | Deposit date: | 2020-06-08 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The interaction between RPAP3 and TRBP reveals a possible involvement of the HSP90/R2TP chaperone complex in the regulation of miRNA activity.
Nucleic Acids Res., 50, 2022
|
|
1KIX
 
 | |
1KO5
 
 | | Crystal structure of gluconate kinase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Gluconate kinase, MAGNESIUM ION | | Authors: | Kraft, L, Sprenger, G.A, Lindqvist, Y. | | Deposit date: | 2001-12-20 | | Release date: | 2002-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Conformational changes during the catalytic cycle of gluconate kinase as revealed by X-ray crystallography.
J.Mol.Biol., 318, 2002
|
|
6ZHL
 
 | | Crystal Structure of Staphylococcus aureus RsgA bound to ppGpp. | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE-5',3'-TETRAPHOSPHATE, Small ribosomal subunit biogenesis GTPase RsgA, ... | | Authors: | Bennison, D.J, Rafferty, J.B, Corrigan, R.M. | | Deposit date: | 2020-06-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The Stringent Response Inhibits 70S Ribosome Formation in Staphylococcus aureus by Impeding GTPase-Ribosome Interactions.
Mbio, 12, 2021
|
|
6Z5Y
 
 | |
5BU6
 
 | | Structure of BpsB deaceylase domain from Bordetella bronchiseptica | | Descriptor: | 1,2-ETHANEDIOL, BpsB (PgaB), Poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase, ... | | Authors: | Little, D.J, Bamford, N.C, Howell, P.L. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | The Protein BpsB Is a Poly-beta-1,6-N-acetyl-d-glucosamine Deacetylase Required for Biofilm Formation in Bordetella bronchiseptica.
J.Biol.Chem., 290, 2015
|
|
6YVM
 
 | | Human OMPD-domain of UMPS in complex with the substrate OMP at 1.25 Angstroms resolution, 2.13 MGy exposure | | Descriptor: | GLYCEROL, OROTIDINE-5'-MONOPHOSPHATE, SULFATE ION, ... | | Authors: | Tittmann, K, Rindfleisch, S, Krull, M. | | Deposit date: | 2020-04-28 | | Release date: | 2021-11-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ground-state destabilization by electrostatic repulsion is not a driving force in orotidine-5-monophosphate decarboxylase catalysis
Nat Catal, 5, 2022
|
|
6C9R
 
 | | Mycobacterium tuberculosis adenosine kinase bound to (2R,3S,4R,5R)-2-(hydroxymethyl)-5-(6-(thiophen-3-yl)-9H-purin-9-yl)tetrahydrofuran-3,4-diol | | Descriptor: | 9-beta-D-ribofuranosyl-6-(thiophen-3-yl)-9H-purine, Adenosine kinase, GLYCEROL, ... | | Authors: | Crespo, R.A, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2018-01-28 | | Release date: | 2019-05-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Drug Design of 6-Substituted Adenosine Analogues as Potent Inhibitors of Mycobacterium tuberculosis Adenosine Kinase.
J.Med.Chem., 62, 2019
|
|
