2VQQ
 
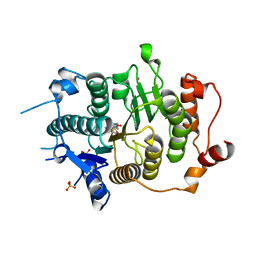 | | Structure of HDAC4 catalytic domain (a double cysteine-to-alanine mutant) bound to a trifluoromethylketone inhbitor | | Descriptor: | 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
4ZHW
 
 | |
2VQJ
 
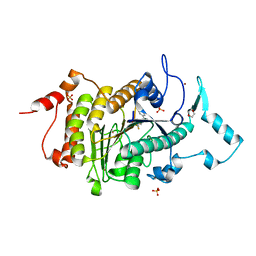 | | Structure of HDAC4 catalytic domain bound to a trifluoromethylketone inhbitor | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2,2,2-TRIFLUORO-1-{5-[(3-PHENYL-5,6-DIHYDROIMIDAZO[1,2-A]PYRAZIN-7(8H)-YL)CARBONYL]THIOPHEN-2-YL}ETHANE-1,1-DIOL, HISTONE DEACETYLASE 4, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2W57
 
 | |
3GA1
 
 | |
2Z9O
 
 | |
4DOS
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to DLPC and a Fragment of TIF-2 | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Musille, P.M, Ortlund, E.A. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2XJ3
 
 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
3F8Z
 
 | | Human Dihydrofolate Reductase Structural Data with Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2008-11-13 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
2VQW
 
 | | Structure of inhibitor-free HDAC4 catalytic domain (with gain-of- function mutation His332Tyr) | | Descriptor: | HISTONE DEACETYLASE 4, POTASSIUM ION, ZINC ION | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
2VQM
 
 | | Structure of HDAC4 catalytic domain bound to a hydroxamic acid inhbitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-17 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
3FS6
 
 | | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes | | Descriptor: | 2,4-DIAMINO-5-[2-METHOXY-5-(4-CARBOXYBUTYLOXY)BENZYL]PYRIMIDINE, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cody, V, Pace, J, Makin, J, Piraino, J, Queener, S.F, Rosowsky, A. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Correlations of Inhibitor Kinetics for Pneumocystis jirovecii and Human Dihydrofolate Reductase with Structural Data for Human Active Site Mutant Enzyme Complexes.
Biochemistry, 48, 2009
|
|
3GLB
 
 | | Crystal structure of the effector binding domain of a CATM variant (R156H) | | Descriptor: | (2Z,4Z)-HEXA-2,4-DIENEDIOIC ACID, GLYCEROL, HTH-type transcriptional regulator catM, ... | | Authors: | Ezezika, O.C, Craven, S.H, Neidle, E.L, Momany, C. | | Deposit date: | 2009-03-11 | | Release date: | 2009-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Inducer responses of BenM, a LysR-type transcriptional regulator from Acinetobacter baylyi ADP1.
Mol.Microbiol., 72, 2009
|
|
1PD7
 
 | | Extended SID of Mad1 bound to the PAH2 domain of mSin3B | | Descriptor: | Mad1, Sin3b protein | | Authors: | Van Ingen, H, Lasonder, E, Jansen, J.F, Kaan, A.M, Spronk, C.A, Stunnenberg, H.G, Vuister, G.W. | | Deposit date: | 2003-05-19 | | Release date: | 2004-01-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Extension of the binding motif of the sin3 interacting domain of the mad family proteins(,).
Biochemistry, 43, 2004
|
|
6FBZ
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum in the cap-bound state | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E-like protein,Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.496 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6FC3
 
 | |
1I2A
 
 | |
5UC1
 
 | | Structural Analysis of Glucocorticoid Receptor beta Ligand Binding Domain Complexed with Glucocorticoid Antagonist RU-486: Implication of Helix 12 in Antagonism | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, ... | | Authors: | Pedersen, L.C, Min, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Probing Dominant Negative Behavior of Glucocorticoid Receptor beta through a Hybrid Structural and Biochemical Approach.
Mol. Cell. Biol., 2018
|
|
6FC0
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum | | Descriptor: | Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6FC1
 
 | |
7CDC
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRP peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDF
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRK peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDD
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDG
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRRR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
7CDE
 
 | | Crystal structure of LSD1-CoREST in complex with PRSFLVRKR peptide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Lysine-specific histone demethylase 1A, ... | | Authors: | Kikuchi, M, Kitagawa, H, Sato, S, Umezawa, N, Higuchi, T, Umehara, T. | | Deposit date: | 2020-06-19 | | Release date: | 2021-06-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure-Based Identification of Potent Lysine-Specific Demethylase 1 Inhibitor Peptides and Temporary Cyclization to Enhance Proteolytic Stability and Cell Growth-Inhibitory Activity.
J.Med.Chem., 64, 2021
|
|
