1NQS
 
 | | Structural Characterisation of the Holliday Junction formed by the sequence d(TCGGTACCGA) at 1.97 A | | Descriptor: | 5'-d(TpCpGpGpTpApCpCpGpA)-3', CALCIUM ION | | Authors: | Cardin, C.J, Gale, B.C, Thorpe, J.H, Texieira, S.C.M, Gan, Y, Moraes, M.I.A.A, Brogden, A.L. | | Deposit date: | 2003-01-22 | | Release date: | 2003-02-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Analysis of two Holliday junctions formed by the sequences TCGGTACCGA and CCGGTACCGG
To be Published
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1NQU
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 6,7-dioxo-5H-8-ribitylaminolumazine | | Descriptor: | 6,7-DIOXO-5H-8-RIBITYLAMINOLUMAZINE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQV
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-nitroso-6-ribityl-amino-2,4(1H,3H)pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQW
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 5-(6-D-ribitylamino-2,4(1H,3H)pyrimidinedione-5-yl)-1-pentyl-phosphonic acid | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4(1H,3H)PYRIMIDINEDIONE-5-YL) PENTYL-1-PHOSPHONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQX
 
 | | Crystal Structure of Lumazine Synthase from Aquifex aeolicus in Complex with Inhibitor: 3-(7-hydroxy-8-ribityllumazine-6-yl)propionic acid | | Descriptor: | 3-(7-HYDROXY-8-RIBITYLLUMAZINE-6-YL) PROPIONIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Zhang, X, Meining, W, Cushman, M, Haase, I, Fischer, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2003-01-23 | | Release date: | 2004-01-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A structure-based model of the reaction catalyzed by lumazine synthase from Aquifex aeolicus.
J.Mol.Biol., 328, 2003
|
|
1NQY
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein) | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NQZ
 
 | | The structure of a CoA pyrophosphatase from D. Radiodurans complexed with a magnesium ion | | Descriptor: | CoA pyrophosphatase (MutT/nudix family protein), MAGNESIUM ION | | Authors: | Kang, L.W, Gabelli, S.B, Bianchet, M.A, Xu, W.L, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a coenzyme A pyrophosphatase from Deinococcus radiodurans: a member of the Nudix family.
J.Bacteriol., 185, 2003
|
|
1NR0
 
 | | Two Seven-Bladed Beta-Propeller Domains Revealed By The Structure Of A C. elegans Homologue Of Yeast Actin Interacting Protein 1 (AIP1). | | Descriptor: | Actin interacting protein 1, MANGANESE (II) ION | | Authors: | Vorobiev, S.M, Mohri, K, Ono, S, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2003-01-23 | | Release date: | 2003-07-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification of Functional Residues on Caenorhabditis elegans Actin-interacting Protein 1 (UNC-78) for Disassembly of Actin Depolymerizing Factor/Cofilin-bound Actin Filaments
J.Biol.Chem., 279, 2004
|
|
1NR1
 
 | | Crystal structure of the R463A mutant of human Glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1NR2
 
 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NR3
 
 | | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212 | | Descriptor: | DNA-binding protein tfx | | Authors: | Shen, Y, Liu, G, Bhaskaran, R, Yee, A, Arrowsmith, C, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2003-01-23 | | Release date: | 2003-06-10 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | SOLUTION STRUCTURE OF THE PROTEIN MTH0916: THE NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET TT212
To be Published
|
|
1NR4
 
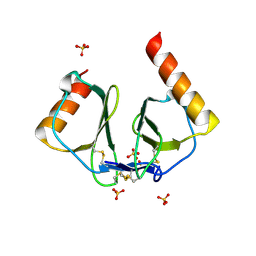 | | High resolution crystal structures of thymus and activation-regulated chemokine | | Descriptor: | SULFATE ION, Thymus and activation-regulated chemokine | | Authors: | Asojo, O.A, Boulegue, C, Hoover, D.M, Lu, W, Lubkowski, J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of thymus and activation-regulated chemokine (TARC).
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1NR5
 
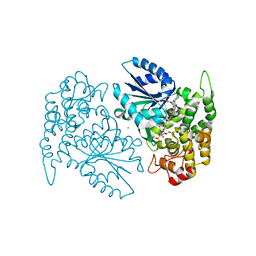 | | Crystal structure of 3-dehydroquinate synthase (DHQS) in complex with ZN2+, NAD and carbaphosphonate | | Descriptor: | 3-DEHYDROQUINATE SYNTHASE, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Nichols, C.E, Ren, J, Lamb, H.K, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2003-01-23 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced Conformational Changes and a Mechanism for Domain Closure in Aspergillus nidulans Dehydroquinate Synthase
J.MOL.BIOL., 327, 2003
|
|
1NR6
 
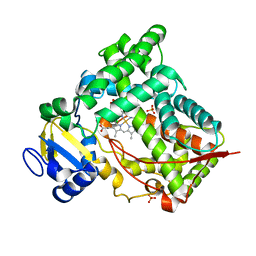 | | MICROSOMAL CYTOCHROME P450 2C5/3LVDH COMPLEX WITH DICLOFENAC | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, CYTOCHROME P450 2C5, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wester, M.R, Johnson, E.F, Stout, C.D. | | Deposit date: | 2003-01-23 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Mammalian Cytochrome P450 2C5 Complexed with Diclofenac at 2.1 A Resolution: Evidence for an Induced Fit Model of Substrate Binding
Biochemistry, 42, 2003
|
|
1NR7
 
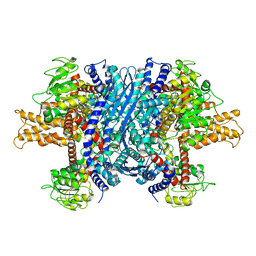 | | Crystal structure of apo bovine glutamate dehydrogenase | | Descriptor: | Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
1NR8
 
 | | The crystal structure of a D-Lysine-based chiral PNA-DNA duplex | | Descriptor: | 5'-D(P*AP*GP*TP*GP*AP*TP*CP*TP*AP*C)-3', H-((GPN)*(TPN)*(APN)*(GPN)*(A66)*(T66)*(C66)*(APN)*(CPN)*(TPN))-NH2, MAGNESIUM ION | | Authors: | Menchise, V, De Simone, G, Tedeschi, T, Corradini, R, Sforza, S, Marchelli, R, Capasso, D, Saviano, M, Pedone, C. | | Deposit date: | 2003-01-24 | | Release date: | 2003-10-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Insights into peptide nucleic acid (PNA) structural features: The
crystal structure of a D-lysine-based chiral PNA-DNA duplex
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NR9
 
 | | Crystal Structure of Escherichia coli 1262 (APC5008), Putative Isomerase | | Descriptor: | MAGNESIUM ION, Protein YCGM | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Skarina, T, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Escherichia coli Putative Isomerase EC1262 (APC5008)
To be Published
|
|
1NRA
 
 | |
1NRB
 
 | |
1NRE
 
 | |
1NRF
 
 | | C-terminal domain of the Bacillus licheniformis BlaR penicillin-receptor | | Descriptor: | REGULATORY PROTEIN BLAR1 | | Authors: | Kerff, F, Charlier, P, Columbo, M.L, Sauvage, E, Brans, A, Frere, J.M, Joris, B, Fonze, E. | | Deposit date: | 2003-01-24 | | Release date: | 2004-01-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the sensor domain of the BlaR penicillin receptor from Bacillus licheniformis.
Biochemistry, 42, 2003
|
|
1NRG
 
 | | Structure and Properties of Recombinant Human Pyridoxine-5'-Phosphate Oxidase | | Descriptor: | BETA-MERCAPTOETHANOL, FLAVIN MONONUCLEOTIDE, PHOSPHATE ION, ... | | Authors: | Musayev, F.N, di Salvo, M.L, Ko, T.P, Schirch, V, Safo, M.K. | | Deposit date: | 2003-01-24 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure and properties of recombinant human pyridoxine 5'-phosphate oxidase.
Protein Sci., 12, 2003
|
|
1NRI
 
 | | Crystal Structure of Putative Phosphosugar Isomerase HI0754 from Haemophilus influenzae | | Descriptor: | Hypothetical protein HI0754 | | Authors: | Kim, Y, Quartey, P, Ng, R, Zarembinski, T.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-24 | | Release date: | 2003-07-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Hypothetical protein HI0754 from Haemophilus influenzae
To be Published
|
|
1NRJ
 
 | |
