2VGV
 
 | | Crystal structure of E53QbsSHMT obtained in the presence of L-allo- Threonine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Rajaram, V, Bhavani, B.S, Kaul, P, Prakash, V, Appaji Rao, N, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2007-11-15 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure Determination and Biochemical Studies on Bacillus Stearothermophilus E53Q Serine Hydroxymethyltransferase and its Complexes Provide Insights on Function and Enzyme Memory
FEBS J., 274, 2007
|
|
3E0V
 
 | |
3E0W
 
 | |
3GRB
 
 | | Crystal structure of the F87M/L110M mutant of human transthyretin at pH 6.5 | | Descriptor: | ACETATE ION, GLYCEROL, Transthyretin, ... | | Authors: | Palmieri, L.C, Freire, J.B.B, Foguel, D, Lima, L.M.T.R. | | Deposit date: | 2009-03-25 | | Release date: | 2010-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel Zn2+-binding sites in human transthyretin: implications for amyloidogenesis and retinol-binding protein recognition.
J.Biol.Chem., 285, 2010
|
|
4OC8
 
 | | DNA modification-dependent restriction endonuclease AspBHI | | Descriptor: | PHOSPHATE ION, restriction endonuclease AspBHI | | Authors: | Horton, J.R. | | Deposit date: | 2014-01-08 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Structure and mutagenesis of the DNA modification-dependent restriction endonuclease AspBHI.
Sci Rep, 4, 2014
|
|
1PYD
 
 | |
7MQM
 
 | | AAC(3)-IIIa in complex with CoA and gentamicin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase III, COENZYME A, GLYCEROL, ... | | Authors: | Zielinski, M, Berghuis, A.M. | | Deposit date: | 2021-05-05 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of substrate-bound aminoglycoside acetyltransferase (3)-IIIa.
Plos One, 17, 2022
|
|
4KAT
 
 | | Crystal structure of FDTS from T. maritima mutant (R174K) with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE-5'-MONOPHOSPHATE, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, Thymidylate synthase | | Authors: | Mathews, I.I. | | Deposit date: | 2013-04-22 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Flavin-Dependent Thymidylate Synthase as a Drug Target for Deadly Microbes: Mutational Study and a Strategy for Inhibitor Design.
J Bioterror Biodef, Suppl 12, 2013
|
|
3CWW
 
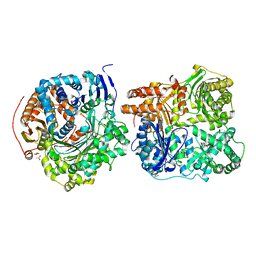 | | Crystal Structure of IDE-bradykinin complex | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETATE ION, Insulin-degrading enzyme, ... | | Authors: | Malito, E, Tang, W.J. | | Deposit date: | 2008-04-23 | | Release date: | 2008-11-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Molecular Bases for the Recognition of Short Peptide Substrates and Cysteine-Directed Modifications of Human Insulin-Degrading Enzyme
Biochemistry, 47, 2008
|
|
8IAH
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAI
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State II, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
3CUS
 
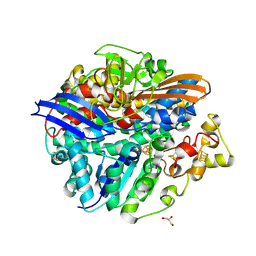 | |
2YXG
 
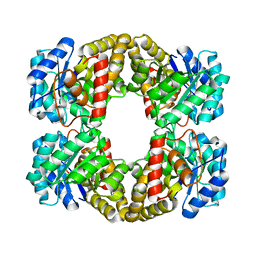 | |
3CKE
 
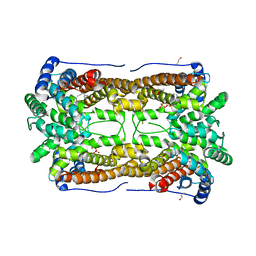 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
8IQ8
 
 | | Crystal structure of 3,4-dihydroxyphenylacetate 2,3-dioxygenase (DHPAO) from Acinetobacter baumannii | | Descriptor: | 3,4-dihydroxyphenylacetate 2,3-dioxygenase, SODIUM ION | | Authors: | Chitnumsub, P, Maenpuen, S. | | Deposit date: | 2023-03-16 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and biochemical characterization of an extradiol 3,4-dihydroxyphenylacetate 2,3-dioxygenase from Acinetobacter baumannii.
Arch.Biochem.Biophys., 747, 2023
|
|
3THO
 
 | |
6F1C
 
 | | C1rC1s complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1r subcomponent, ... | | Authors: | Almitairi, J.O.M, Venkatraman Girija, U, Furze, C.M, Simpson-Gray, X, Badakshi, F, Marshall, J.E, Mitchell, D.A, Moody, P.C.E, Wallis, R. | | Deposit date: | 2017-11-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the C1r-C1s interaction of the C1 complex of complement activation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4QYJ
 
 | |
6PIH
 
 | | Hexameric ArnA cryo-EM structure | | Descriptor: | Bifunctional polymyxin resistance protein ArnA, UDP-4-amino-4-deoxy-L-arabinose formyltransferase | | Authors: | Yang, M, Gehring, K. | | Deposit date: | 2019-06-26 | | Release date: | 2019-07-31 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Cryo-electron microscopy structures of ArnA, a key enzyme for polymyxin resistance, revealed unexpected oligomerizations and domain movements.
J.Struct.Biol., 208, 2019
|
|
6FGJ
 
 | | Crystal structure of the small alarmone synthethase 2 from Staphylococcus aureus | | Descriptor: | GTP pyrophosphokinase, TRIETHYLENE GLYCOL | | Authors: | Bange, G, Steinchen, W, Vogt, M, Altegoer, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structural and mechanistic divergence of the small (p)ppGpp synthetases RelP and RelQ.
Sci Rep, 8, 2018
|
|
6QAK
 
 | |
6G3D
 
 | | Crystal structure of Native EDDS lyase | | Descriptor: | Argininosuccinate lyase | | Authors: | Poddar, H, Thunnissem, A.M.W.H, Poelarends, G.J. | | Deposit date: | 2018-03-25 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.221 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of Ethylenediamine- N, N'-disuccinic Acid Lyase, a Carbon-Nitrogen Bond-Forming Enzyme with a Broad Substrate Scope.
Biochemistry, 57, 2018
|
|
6QAP
 
 | |
3BG5
 
 | | Crystal Structure of Staphylococcus Aureus Pyruvate Carboxylase | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Xiang, S, Tong, L. | | Deposit date: | 2007-11-26 | | Release date: | 2008-02-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of human and Staphylococcus aureus pyruvate carboxylase and molecular insights into the carboxyltransfer reaction.
Nat.Struct.Mol.Biol., 15, 2008
|
|
