7YVV
 
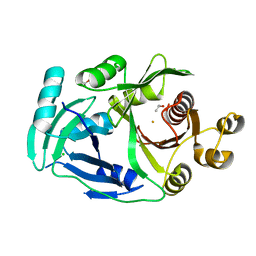 | | AcmP1, R-4-hydroxymandelate synthase | | Descriptor: | 4-hydroxyphenylpyruvate dioxygenase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Zhang, B, Ge, H.M. | | Deposit date: | 2022-08-19 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of AcmP1 as the native R-4-hydroxymandelate synthase from biosynthetic pathway of Amycolamycins
To Be Published
|
|
7MX8
 
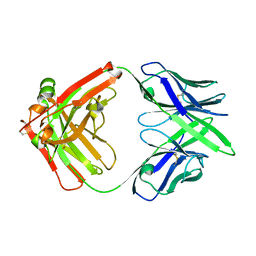 | |
7MY5
 
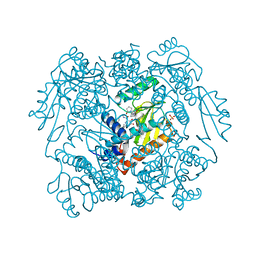 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-2-(2-methylphenyl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988503
To Be Published
|
|
6NFZ
 
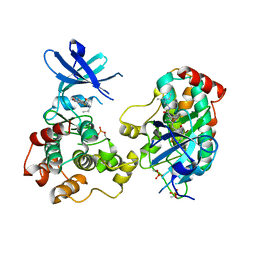 | | Crystal structure of diphosphorylated HPK1 kinase domain in complex with sunitinib in the active state. | | Descriptor: | Mitogen-activated protein kinase kinase kinase kinase 1, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Johnson, E, McTigue, M, Cronin, C.N. | | Deposit date: | 2018-12-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.966 Å) | | Cite: | Multiple conformational states of the HPK1 kinase domain in complex with sunitinib reveal the structural changes accompanying HPK1 trans-regulation.
J.Biol.Chem., 294, 2019
|
|
7PJN
 
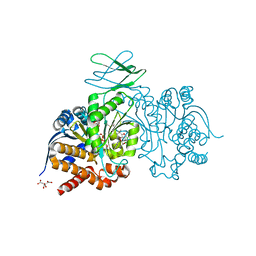 | | Crystal Structure of Ivosidenib-resistant IDH1 variant R132C S280F in complex with NADPH and inhibitor DS-1001B | | Descriptor: | (E)-3-(1-(5-(2-fluoropropan-2-yl)-3-(2,4,6-trichlorophenyl)isoxazole-4-carbonyl)-3-methyl-1H-indol-4-yl)acrylic acid, CITRIC ACID, GLYCEROL, ... | | Authors: | Reinbold, R, Rabe, P, Abboud, M.I, Schofield, C.J, Clifton, I.J. | | Deposit date: | 2021-08-24 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Resistance to the isocitrate dehydrogenase 1 mutant inhibitor ivosidenib can be overcome by alternative dimer-interface binding inhibitors.
Nat Commun, 13, 2022
|
|
6NG2
 
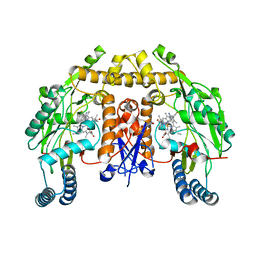 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-(3-(dimethylamino)prop-1-yn-1-yl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)prop-1-yn-1-yl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
8PX2
 
 | |
6NGD
 
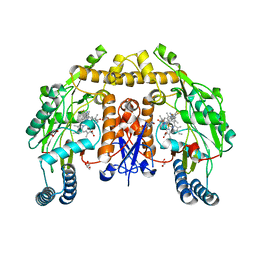 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(5-(3-(dimethylamino)propyl)-2,3,4-trifluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[3-(dimethylamino)propyl]-2,3,4-trifluorophenyl}ethyl)-4-methylpyridin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
5DOI
 
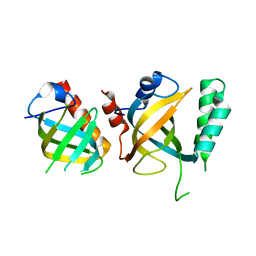 | | Crystal structure of Tetrahymena p45N and p19 | | Descriptor: | Telomerase associated protein p45, Telomerase-associated protein 19 | | Authors: | Wan, B, Tang, T, Wu, J, Lei, M. | | Deposit date: | 2015-09-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Tetrahymena telomerase p75-p45-p19 subcomplex is a unique CST complex
Nat.Struct.Mol.Biol., 22, 2015
|
|
8PPN
 
 | |
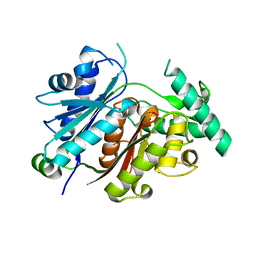
7YVY
 
 | |
8SOQ
 
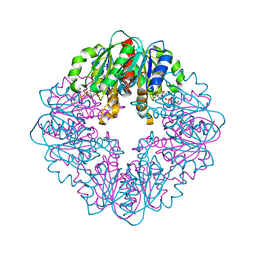 | | S127A variant of LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with authentic substrate NaAD | | Descriptor: | MAGNESIUM ION, NICOTINIC ACID ADENINE DINUCLEOTIDE, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase | | Authors: | Chatterjee, S, Rankin, J.A, Hu, J, Hausinger, R.P. | | Deposit date: | 2023-04-29 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the LarB-Substrate Complex and Identification of a Reaction Intermediate during Nickel-Pincer Nucleotide Cofactor Biosynthesis.
Biochemistry, 62, 2023
|
|
7MY1
 
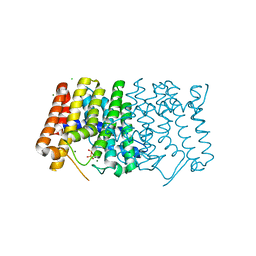 | | Sy-CrtE structure with IPP, N-term His-tag | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, ... | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
5HQG
 
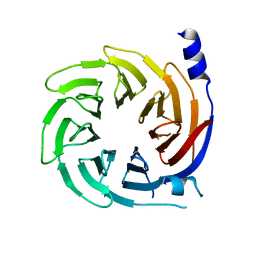 | |
7UP2
 
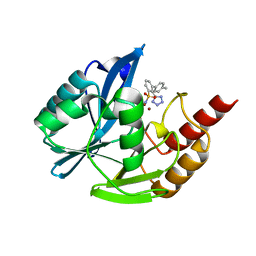 | | NDM1-inhibitor co-structure | | Descriptor: | (2M)-4'-methyl-2-(2H-tetrazol-5-yl)[1,1'-biphenyl]-3-sulfonamide, Beta-lactamase VIM-1, ZINC ION | | Authors: | Scapin, G, Fischmann, T.O. | | Deposit date: | 2022-04-14 | | Release date: | 2023-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Rapid Evolution of a Fragment-like Molecule to Pan-Metallo-Beta-Lactamase Inhibitors: Initial Leads toward Clinical Candidates.
J.Med.Chem., 65, 2022
|
|
6NGR
 
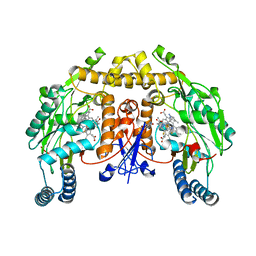 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(2-((2R,4S)-4-fluoro-1-methylpyrrolidin-2-yl)ethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[2-(3-fluoro-5-{2-[(2R,4S)-4-fluoro-1-methylpyrrolidin-2-yl]ethyl}phenyl)ethyl]-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7YUJ
 
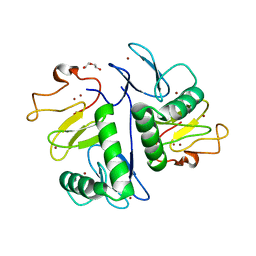 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
5VDR
 
 | |
7MXZ
 
 | | Sy-CrtE apo structure | | Descriptor: | CHLORIDE ION, Geranylgeranyl pyrophosphate synthase, MAGNESIUM ION | | Authors: | Peat, T.S, Newman, J. | | Deposit date: | 2021-05-19 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Molecular characterization of cyanobacterial short-chain prenyltransferases and discovery of a novel GGPP phosphatase.
Febs J., 289, 2022
|
|
6NH1
 
 | | Structure of human endothelial nitric oxide synthase heme domain in complex with 6-(3-fluoro-5-(3-(methylamino)prop-1-yn-1-yl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-fluoro-5-[3-(methylamino)prop-1-yn-1-yl]phenyl}ethyl)-4-methylpyridin-2-amine, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-03-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.216 Å) | | Cite: | Optimization of Blood-Brain Barrier Permeability with Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibitors Having a 2-Aminopyridine Scaffold.
J. Med. Chem., 62, 2019
|
|
7YUI
 
 | |
8PCW
 
 | | Structure of Csm6' from Streptococcus thermophilus | | Descriptor: | CRISPR system endoribonuclease Csm6' | | Authors: | McQuarrie, S.J, Athukoralage, J.S, McMahon, S.A, Graham, S, Ackerman, K, Bode, B.E, White, M.F, Gloster, T.M. | | Deposit date: | 2023-06-11 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.54 Å) | | Cite: | Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.
Nucleic Acids Res., 51, 2023
|
|
5E7W
 
 | | X-ray Structure of Human Recombinant 2Zn insulin at 0.92 Angstrom | | Descriptor: | ACETATE ION, Insulin, N-PROPANOL, ... | | Authors: | Lisgarten, D.R, Naylor, C.E, Palmer, R.A, Lobley, C.M.C. | | Deposit date: | 2015-10-13 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.9519 Å) | | Cite: | Ultra-high resolution X-ray structures of two forms of human recombinant insulin at 100 K.
Chem Cent J, 11, 2017
|
|
7UKA
 
 | |
8SPE
 
 | | Crystal structure of Bax core domain BH3-groove dimer - tetrameric fraction P31 | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BAX, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Miller, M.S, Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2023-05-03 | | Release date: | 2023-12-27 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence differences between BAX and BAK core domains manifest as differences in their interactions with lipids.
Febs J., 291, 2024
|
|
